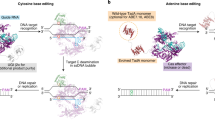
Abstract
Base editing is a powerful CRISPR-based technology for introducing precise substitutions into the genome. This technology greatly advances mutagenesis possibilities in vivo, particularly in zebrafish, for which the generation of precise point mutations is still challenging. Zebrafish have emerged as an important model for genetic studies and in vivo disease modeling. With the development of different base editor variants that recognize protospacer-adjacent motifs (PAMs) other than the classical 5′-NGG-3′ PAM, it is now possible to design and test several guide RNAs to find the most efficient way to precisely introduce the desired substitution. Here, we describe the experimental design strategies and protocols for cytosine base editing in zebrafish, from guide RNA design and selection of base editor variants to generation of the zebrafish mutant line carrying the substitution of interest. By using co-selection by introducing a loss-of-function mutation in genes necessary for the formation of pigments, injected embryos with highly efficient base editing can be directly analyzed to determine the phenotypic impact of the targeted substitution. The generation of mutant embryos after base editor injections in zebrafish can be completed within 2 weeks.
Key points
-
Cytosine base editing can introduce specific C-to-T transitions in the genome. This protocol for cytosine base editing in zebrafish covers guide RNA design, selection of base editors and generation of mutant lines carrying the substitution of interest.
-
Compared with standard CRISPR–Cas9-based approaches, base editors generate precise substitutions without double-strand DNA breaks, improving efficiency over previous methods to generate zebrafish lines with precise human pathological mutations for in vivo disease modeling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018).
Rosello, M. et al. Disease modeling by efficient genome editing using a near PAM-less base editor in vivo. Nat. Commun. 13, 3435 (2022).
Rosello, M. et al. Precise base editing for the in vivo study of developmental signaling and human pathologies in zebrafish. eLife 10, e65552 (2021).
Zhao, Y., Shang, D., Ying, R., Cheng, H. & Zhou, R. An optimized base editor with efficient C-to-T base editing in zebrafish. BMC Biol. 18, 190 (2020).
Qin, W. et al. Precise A•T to G•C base editing in the zebrafish genome. BMC Biol. 16, 139 (2018).
Liang, F. et al. SpG and SpRY variants expand the CRISPR toolbox for genome editing in zebrafish. Nat. Commun. 13, 3421 (2022).
Cornean, A. et al. Precise in vivo functional analysis of DNA variants with base editing using ACEofBASEs target prediction. elife 11, e72124 (2022).
Kantor, A., McClements, M. E. & MacLaren, R. E. CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21, 6240 (2020).
Zeballos, C. M. & Gaj, T. Next-generation CRISPR technologies and their applications in gene and cell therapy. Trends Biotechnol. 39, 692–705 (2021).
Porto, E. M., Komor, A. C., Slaymaker, I. M. & Yeo, G. W. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19, 839–859 (2020).
Sasaguri, H. et al. Introduction of pathogenic mutations into the mouse Psen1 gene by Base Editor and Target-AID. Nat. Commun. 9, 2892 (2018).
Wang, F. et al. Generation of a Hutchinson-Gilford progeria syndrome monkey model by base editing. Protein Cell 11, 809–824 (2020).
Carreras, A. et al. In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol. 17, 4 (2019).
Zhang, H. et al. Adenine base editing in vivo with a single adeno-associated virus vector. GEN Biotechnol. 1, 285–299 (2022).
Xie, J. et al. Efficient base editing for multiple genes and loci in pigs using base editors. Nat. Commun. 10, 2852 (2019).
Musunuru, K. et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021).
Santoriello, C. & Zon, L. I. Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 122, 2337–2343 (2012).
Bradford, Y. M. et al. Zebrafish models of human disease: gaining insight into human disease at ZFIN. ILAR J. 58, 4–16 (2017).
Patton, E. E. & Tobin, D. M. Spotlight on zebrafish: the next wave of translational research. Dis. Model. Mech. 12, dmm039370 (2019).
Adamson, K. I., Sheridan, E. & Grierson, A. J. Use of zebrafish models to investigate rare human disease. J. Med. Genet. 55, 641–649 (2018).
Mione, M. C. & Trede, N. S. The zebrafish as a model for cancer. Dis. Model. Mech. 3, 517–523 (2010).
Schulze, L. et al. Transparent Danionella translucida as a genetically tractable vertebrate brain model. Nat. Methods 15, 977–983 (2018).
Park, D. S. et al. Targeted base editing via RNA-guided cytidine deaminases in Xenopus laevis embryos. Mol. Cells 40, 823–827 (2017).
Shi, Z. et al. Modeling human point mutation diseases in Xenopus tropicalis with a modified CRISPR/Cas9 system. FASEB J. 33, 6962–6968 (2019).
Li, S. et al. Universal toxin-based selection for precise genome engineering in human cells. Nat. Commun. 12, 497 (2021).
Zhao, D. et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40 (2021).
Chen, L. et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 12, 1384 (2021).
Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46 (2021).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Petri, K. et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 40, 189–193 (2022).
Armstrong, G. A. et al. Homology directed knockin of point mutations in the Zebrafish tardbp and fus genes in ALS using the CRISPR/Cas9 system. PloS One 11, e0150188 (2016).
Irion, U., Krauss, J. & Nusslein-Volhard, C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141, 4827–4830 (2014).
Zhang, Y., Huang, H., Zhang, B. & Lin, S. TALEN- and CRISPR-enhanced DNA homologous recombination for gene editing in zebrafish. Methods Cell Biol. 135, 107–120 (2016).
Prykhozhij, S. V. et al. Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res. 46, e102 (2018).
Albadri, S., Del Bene, F. & Revenu, C. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121–122, 77–85 (2017).
Hoshijima, K., Jurynec, M. J. & Grunwald, D. J. Precise editing of the zebrafish genome made simple and efficient. Dev. Cell 36, 654–667 (2016).
Wierson, W. A. et al. Efficient targeted integration directed by short homology in zebrafish and mammalian cells. eLife 9, e53968 (2020).
Burger, A. et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025–2037 (2016).
Shah, A. N., Davey, C. F., Whitebirch, A. C., Miller, A. C. & Moens, C. B. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods 12, 535–540 (2015).
Wu, R. S. et al. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125.e4 (2018).
Dooley, C. M. et al. Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Melanoma Res. 26, 205–217 (2013).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Grunewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019).
Rembold, M., Lahiri, K., Foulkes, N. S. & Wittbrodt, J. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. Nat. Protoc. 1, 1133–1139 (2006).
Kluesner, M. G. et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018).
Xing, L., Quist, T. S., Stevenson, T. J., Dahlem, T. J. & Bonkowsky, J. L. Rapid and efficient zebrafish genotyping using PCR with high-resolution melt analysis. J. Vis. Exp. 2014, e51138 (2014).
Acknowledgements
We thank C. Giovannangeli and A. Miccio for helpful discussions and sharing plasmids. M.R. was supported by the Fondation pour la Recherche Médicale (FRM grant number ECO20170637481) and la Ligue Nationale Contre le Cancer. This work was supported by ANR-18-CE16 ‘iReelAx’, ANR-20-CE17-0020-02 ‘INCEPTION’, ANR-11-INBS-0014-TEFOR, UNADEV in partnership with ITMO NNP/AVIESAN (national alliance for life sciences and health, UNADEV-19UU51-DEL BENE), Fondation pour la Recherche Médicale (MND202003011485), in the framework of research on vision and IHU FOReSIGHT (ANR-18-IAHU-0001) supported by French state funds managed by the Agence Nationale de la Recherche within the Investissements d’Avenir program.
Author information
Authors and Affiliations
Contributions
All the authors contributed to the protocol development. M.R. and M.S. did the experimental work. M.R. and F.D.B. wrote the manuscript with input and editing from J.-P.C. and M.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Stephen Ekker and the other, anonymous, reviewers(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Rosello, M. et al. Nat. Commun. 13, 3435 (2022): https://doi.org/10.1038/s41467-022-31172-z
Rosello, M. et al. eLlife 10, e65552 (2021): https://doi.org/10.7554/eLife.65552
Source data
Source Data Fig. 5
Unprocessed gels
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rosello, M., Serafini, M., Concordet, JP. et al. Precise mutagenesis in zebrafish using cytosine base editors. Nat Protoc 18, 2794–2813 (2023). https://doi.org/10.1038/s41596-023-00854-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00854-3
This article is cited by
-
Exploring hematopoiesis in zebrafish using forward genetic screening
Experimental & Molecular Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.