Abstract
The escalating drug resistance among microorganisms underscores the urgent need for innovative therapeutic strategies and a comprehensive understanding of bacteria's defense mechanisms against oxidative stress and antibiotics. Among the recently discovered barriers, the endogenous production of hydrogen sulfide (H2S) via the reverse transsulfuration pathway, emerges as a noteworthy factor. In this study, we have explored the catalytic capabilities and crystal structure of cystathionine γ-lyase from Pseudomonas aeruginosa (PaCGL), a multidrug-opportunistic pathogen chiefly responsible for nosocomial infections. In addition to a canonical l-cystathionine hydrolysis, PaCGL efficiently catalyzes the production of H2S using l-cysteine and/or l-homocysteine as alternative substrates. Comparative analysis with the human enzyme and counterparts from other pathogens revealed distinct structural features within the primary enzyme cavities. Specifically, a distinctly folded entrance loop could potentially modulate the access of substrates and/or inhibitors to the catalytic site. Our findings offer significant insights into the structural evolution of CGL enzymes across different pathogens and provide novel opportunities for developing specific inhibitors targeting PaCGL.
Similar content being viewed by others
Introduction
The diseases caused by bacteria, fungi, or parasites pose a growing problem for today’s healthcare system. Infectious and parasitic diseases are among the top ten global causes of mortality, as identified by the World Health Organization1. Of particular concern is the opportunistic Gram-negative bacterium Pseudomonas aeruginosa, which has been designated as a critical priority for study, discovery, and the development of new antibiotics. Infections caused by P. aeruginosa can progress to extensive colonization and be more severe with a higher mortality rate, especially in cystic fibrosis patients and immunocompromised hospitalized individuals2. At present, treating P. aeruginosa infections effectively poses a substantial challenge owing to the bacterium’s increasing resistance to numerous clinically available antibiotics.
Recent studies have shown that a defense mechanism of bacteria against reactive oxygen species (ROS) and antibiotic-induced oxidative damage relies on the endogenous production of hydrogen sulfide (H2S)3,4,5,6. Based on these findings, inhibiting the endogenous generation of this gasotransmitter has been proposed as a strategy to combat these pathogens3,4,7,8,9,10,11,12, although it is still a matter of debate whether the H2S defensive role applies equally to all bacterial species13,14. Therefore, the detailed study and characterization of the enzymes involved in production of H2S in these organisms becomes particularly relevant for a quest to find novel avenues how to fight multidrug resistance including discovery of new antibiotics or potentiation of the current ones.
The synthesis of H2S in bacteria varies across species and depends on substrate availability and presence of the specific enzymes. Under anaerobic conditions, the primary route for H2S production is the sulfate reduction pathway (SRP) involving the reduction of inorganic sulfate (SO42−) to H2S15 (Fig. 1A). However, under aerobic conditions, the production of H2S by the SRP is quite inefficient compared to the specialized sulfate-reducing bacteria. Therefore, some bacteria can synthetize H2S through 3-mercaptopyruvate sulfurtransferase (3-MST) under conditions of sulfur limitation3 (Fig. 1A). Finally, the third known bacterial mechanism to produce H2S is the reverse transsulfuration pathway (RTP), which involves the conversion of l-homocysteine (l-Hcys) to l-cysteine (l-Cys) through two consecutive steps catalyzed by two distinct pyridoxal-5′-phosphate (PLP)-dependent enzymes, the cystathionine β-synthase (CBS) and the cystathionine γ-lyase (CGL) (Fig. 1A). CBS catalyzes a β-replacement reaction in which the hydroxyl group of l-serine (l-Ser) is replaced by l-Hcys, yielding l-cystathionine (l-Cth) and H2O (reviewed in16). Next, CGL catalyzes the α,γ-elimination of l-Cth into l-Cys, α-ketobutyrate, and ammonia (Fig. 1B, reaction 1). In addition, many CGLs can catalyze β-elimination of l-Cth as a side reaction, producing l-Hcys, pyruvate, and ammonia (Fig. 1B, reaction 2). This reaction is referred to as the β-lyase activity of CGL. Besides these canonical reactions, both CBS and CGL can catalyze the synthesis of H2S using l-Cys and l-Hcys as substrates exploiting alternative reactivity (Fig. 1B, reactions 3 to 7).
Bacterial H2S synthesis and reactions catalyzed by CGLs. (A) Scheme of the known H2S production pathways in bacteria: Sulfate Reduction Pathway (SRP) (grey arrows), 3-Mercaptopyruvate Sulfurtransferase (3-MST) pathway (dashed arrows) and Reverse Transsulfuration Pathway (RTP) (black arrows). The first step in the SRP is the activation of sulfate by the bifunctional sulfate adenylyltransferase subunit1/adenylylsulfate kinase (CysND), which converts ATP and sulfate to AMP and adenosine 5′-phosphosulfate (APS). The APS is transformed into 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by adenylyl-sulfate kinase (CysC), and then reduced to sulfite (SO32−) by phosphoadenosine phosphosulfate reductase (CysH). Sulfite is subsequently reduced to hydrogen sulfide (H2S) by sulfite reductase (SR, CysI). On the other hand, the 3-MST pathway initiates with the conversion of cysteine into 3-mercaptopyruvate by the enzyme cystine aminotransferase (CAT). CAT transfers an amino group from cysteine to α-ketoglutarate, yielding 3-mercaptopyruvate and glutamate. Once formed, 3-mercaptopyruvate serves as a substrate for the enzyme 3-Mercaptopyruvate Sulfurtransferase (3-MST). 3-MST catalyzes the transfer of a sulfur atom from 3-mercaptopyruvate to an acceptor molecule, leading to the production of H2S. Finally, the H2S production via RTP (explained detail in the “Introduction” section) involves the desulfuration of cysteine. Key enzymes in RTP are cystathionine β-synthase (CBS)31 and cystathionine γ-lyase (CSE) (widely described in this manuscript). Abbreviations: SO42− (sulfate); SO32− (sulfite); 3-mercaptopyruvate sulfurtransferase (3-MST); persulfidated 3-MST (3-MST-SS); Cystathionine β-synthase (CBS); Cystathionine γ-lyase (CGL); (B) Reactions catalyzed by CGL. The first α,γ-elimination of l-Cth (reaction 1) represents the canonical CGL reaction, which does not lead to production of H2S. The α,β elimination of L-Cth (reaction 2) refers to as the β-lyase activity of CGL which, like reaction 1, does not produce H2S. However, CGL catalyzes several alternative reactions using l-Hcys and/or l-Cys as substrates generating H2S (reactions 3–7).
Recent studies have revealed the presence of the Cse gene within P. aeruginosa genome encoding for the CGL (PaCGL) and that inactivation of the Cse gene leads to a reduction of H2S production in clinical isolates of P. aeruginosa3,4. Furthermore, the studies have demonstrated that experimental inhibitors of human CGL (HsCGL) are much less effective or entirely ineffective targeting homologous bacterial CGLs4. This is likely due to structural and physico-chemical differences among CGLs, which could be exploited to develop new antibiotics and/or adjuvants to treat recurrent infections.
In this study we expressed, purified, and biochemically characterized PaCGL as well as solved its crystal structure. We found that PaCGL hydrolyzes l-Cth via γ- and β-elimination mechanisms (Fig. 1B, reactions 1, 2) and, in addition, catalyzes the generation of H2S from l-Cys or/and l-Hcys (Fig. 1B, reactions 3–7). Structural comparisons to other CGL enzymes and a complementary analysis of PaCGL using deep learning predictions by AlphaFold2 (AF2) revealed significant structural diversity within the main cavities of the enzymes as well as a differently organized entrance loop that may regulate the access of substrates and/or inhibitors into the PLP-containing catalytic center. These findings open new avenues for the design of selective inhibitors of PaCGL and provide significant fundamental insights into the structural evolution of CGL enzymes.
Results
Production and biochemical properties of PaCGL
PaCGL was overexpressed in E. coli and purified to homogeneity using Ni-NTA chromatography (Fig. S1A, Fig. S8). Gel filtration of the purified PaCGL showed a molecular mass of 173 kDa, suggesting that it forms tetramers, in accordance with a subunit size of ~ 44 kDa (Fig. S1B). The UV–Vis absorption spectrum of PaCGL at pH 8.0 exhibited, in addition to the protein band centered at 278 nm, a peak at 425 nm that is typical of the ketoenamine tautomer of the internal aldimine (protein-bound PLP) (Fig. S1C). PaCGL binds ~ 1 mol of PLP/mol of CGL subunit with a Kd value for PLP of 0.15 ± 0.01 µM, as calculated by fluorescence titrations of apo-PaCGL with PLP (Fig. S1D). The apo-PaCGL displayed substantially decreased thermal stability with a melting temperature (Tm) of 57 °C compared to the PLP-loaded holo-enzyme yielding Tm of 68 °C (Fig. S1E).
Enzymatic properties of PaCGL enzyme
Canonical reactions
The reaction catalyzed by CGL in the transsulfuration pathway involves elimination at the γ-carbon of l-Cth. However, it is well established17,18,19 that CGLs, owing to the chemistry of the catalyzed reaction, exhibit an appreciable cystathionine β-lyase (CBL)-like activity, i.e., they can cleave both the C–γ–S and C–β–S bonds of l-Cth producing l-Cys or l-Hcys, respectively (Fig. 1B, reactions 1,2). To distinguish between these two elimination reactions, the reaction products were analyzed by LC–MS/MS. As shown in Fig. 2A, when l-Cth (m/z = 223) was used as a substrate, both l-Cys (m/z = 122) and l-Hcys (m/z = 136) were observed, consistent with the enzyme’s ability to catalyze both γ-elimination (reaction 1) and β-elimination (reaction 2) of l-Cth.
Hydrolysis of l-Cth by recombinant PaCGL. (A) LC–MS/MS chromatogram of the amino acid products of l-Cth hydrolysis by PaCGL. Parent ions with m/z values of 122 (l-Cys), 136 (l-Hcys), and 223 (l-Cth) are seen. (B) Steady-state initial velocity kinetics for PaCGL in the hydrolysis of l-Cth measured using the DTNB assay. (C) Kinetics of the l-Cth hydrolysis by PaCGL followed by measuring the pyruvate production via the LDH assay. Each data point in (B) and (C) represents the mean ± SEM of at least three independent experiments. (D) The UV–Vis absorbance spectra of 15 µM PaCGL in the absence (black line) and presence of 2.5 mM l-Cth (red line). (E) CD spectra of 1 mg mL−1 PaCGL in the absence (black line) and presence of 2.5 mM l-Cth (red line).
The kinetics of l-Cth hydrolysis by PaCGL was characterized using the DTNB assay. Firstly, we determined the optimal pH and temperature for PaCGL activity, which were pH 8 and 42 °C, respectively (Fig. S2). However, considering the physiological temperature of 37 °C, we opted to conduct enzymatic characterization of PaCGL at pH 8.0 and 37 °C. The enzymatic kinetics for the hydrolysis of l-Cth by PaCGL revealed kcat and Km values of 5.2 ± 0.2 s−1 and 0.30 ± 0.04 mM, respectively (Fig. 2B, Table 1). The kinetics of pyruvate formation (reaction 2) from l-Cth was also characterized using the LDH assay (Fig. 2C and Table 1).
We also monitored the behavior of the enzyme-bound PLP in the presence of l-Cth by absorption and CD spectroscopy (Fig. 2D,E). Addition of l-Cth to PaCGL resulted in the pronounced absorption difference in the region of 440–480 nm with positive CD signals. For PLP-dependent enzymes involved in γ- and β-elimination reactions, absorbance bands in the region of 440–480 nm usually are assigned to the α-aminocrotonate or α-aminoacrylate intermediates20,21. The absorption region of 320–340 nm is not considered because pyruvate produced during the turnover also contributes to the absorption in this region.
Alternative reactions
The H2S-producing ability of PaCGL was assessed by examining the various reactions outlined in Fig. 1B (reactions 3–7). The product analysis using LC–MS/MS provided direct evidence for all five potential H2S-generating reactions attributed to CGL. When l-Cys served as a substrate, we observed formation of both l-Ser (m/z = 106) and l-Lanthionine (m/z = 209), confirming the enzyme's ability performing both l-Cys β-lyase (reaction 3) and l-Cys β-replacement (reaction 4) activities, respectively (Fig. 3A). Similarly, in the presence of l-Hcys alone, l-Homoserine (m/z = 120) and l-Homolanthionine (m/z = 237) were detected, consistent with γ-elimination (reaction 5) and γ-replacement (reaction 6) reactions (Fig. 3B). In the presence of both l-Hcys and l-Cys, l-Cth (m/z = 223) was detected, consistent with a replacement reaction (reaction 7) (Fig. 3C). Specifically, product analysis in the presence of 10 mM l-Cys and increasing concentration of l-Hcys confirmed that l-Cth production increases with higher l-Hcys concentrations (Fig. 3D).
H2S alternative reactions. (A–D) Product analysis by LC–MS/MS of the PaCGL-catalyzed reactions in the presence of l-Cys alone (A), l-Hcys alone (B) or l-Cys and l-Hcys (C,D). Parent ions with m/z values of 122 (l-Cys), 106 (l-Ser), 209 (l-Lanthionine), 136 (l-Hcys), 120 (l-Homoserine), 237 (l-Homolanthionine) and 223 (l-Cth) are seen. (E) Kinetics of H2S (reaction 3 + 4, black line) and pyruvate (reaction 3, red line) generation by PaCGL in the presence of l-Cys. Each data point represents the mean ± SEM of at least three independent experiments. (F) Kinetics of H2S generation (reactions 5 + 6) by PaCGL in the presence of l-Hcys.
Next, we determined the kinetics of H2S generation from l-Cys or l-Hcys. The active site pocket of CGL has binding requirements for two amino acids, since the main substrate of the enzyme l-Cth is a condensation product of l-Ser and l-Hcys. In the H2S-generating reactions catalyzed by PaCGL (Fig. 1B, reactions 3–7), either one (reaction 3 and 5) or both (reactions 4, 6, and 7) amino acid binding pockets are occupied. We analyzed the kinetic parameters associated with the single substrate reaction (i.e. ignoring H2O) or bimolecular reactions involving two amino acids, as outlined in the Experimental section.
The dependence of the rate of H2S formation on l-Cys concentration is markedly biphasic, simplifying the deconvolution of kinetic parameters related to two distinct phases corresponding to reactions 3 (β-elimination of l-Cys) and 4 (β-replacement of l-Cys). The enzyme kinetics of pyruvate (reaction 3) and of H2S (reactions 3 + 4) formation from l-Cys are shown in Fig. 3E and the resulting kinetic parameters are detailed in Table 1. Notably, PaCGL exhibits significantly higher affinity for l-Cys at site 1 (0.43 ± 0.03 mM) compared to site 2 (33 ± 6 mM), with observed cooperativity in the binding of the second l-Cys molecule (n = 3 ± 1). Interestingly, like many other CGLs19,22,23,24, PaCGL also displayed inhibition by l-Cys (Table 1 and Fig. 3E).
The rate of H2S formation from l-Hcys is not as markedly biphasic as observed for l-Cys (Fig. 3F), even though LC–MS/MS data clearly indicate that both l-Homoserine (reaction 5) and l-Homolanthionine (reaction 6) are produced when l-Hcys serves as a substrate (Fig. 3B). The kinetic parameters for the overall rate of H2S formation (reaction 5 + 6) are reported in Table 1.
Overall structure of PaCGL
The crystal structure of PaCGL complexed with PLP was solved at 2.0 Å (Fig. 4). The few missing or disordered segments not visible in the crystals were predicted by AF225. As expected, the overall fold of PaCGL is consistent with the type-I PLP-dependent enzymes, resembling human, yeast, and bacteria CGLs, as well as enzymes like cystathionine γ-synthase (CGS), or cystathionine β-lyase (CBL)22,26. Each PaCGL subunit consists of 394 amino acids distributed in three domains: (i) the N-terminal domain (residues 1–62), (ii) the central PLP-binding domain (residues 63–260), and (iii) the C-terminal domain (residues 261–394). The N-terminal domain begins with an unstructured segment (residues 1–13), followed by a short α-helix (α0, residues 14–22) and a long loop (residues L23–60), containing a short helix α1 (residues 55–60) that is visible in only one of the four molecules of the asymmetric unit. In the remaining subunits of the tetramer, the 46–57 region of the loop appears disordered.
Structure of PaCGL. (A) Domain architecture of the PaCGL monomer. (B) Crystal structure of PaCGL. The N-terminal domain (residues 1–62), the PLP-binding domain (residues 63–260), and the C-terminal module (green, residues 261–394) are colored in blue, orange, and green, respectively. PLP is in pink sticks. (C) Structure of PaCGL predicted with AF2. Modelled amino acid residues 1–8, and 46–57 not visible in the crystals, are colored in red. The inset shows the loop L347–370 (in red, and pointed out with an asterisk), which partially defines the entrance to the catalytic cavity, and adopts a markedly different conformation in the crystals than in the predicted PaCGL model. The latter is consistent with the overall structure found in other CGLs (Fig. S7). (D) PaCGL tetrameric organization. The four subunits, A to D, are represented in different surface colors. The tetramer can be interpreted as a dimer of dimers formed by subunits A–B and C–D, respectively. The secondary elements from each subunit are colored according to the domain architecture colors shown in panel (A).
The PLP-binding domain is built up of a seven-stranded, mostly parallel, β-sheet (↑β1↓β7↑β6↑β5↑β4↑β2↑β3), with strand β7 (residues 219–223) antiparallel to the rest. This domain additionally contains eight α-helices, split in two sets (α1, α2, α5, α6, α7 and α3, α4, α8, respectively) that flank the central β-sheet core at both sides. The PLP-binding domain houses the catalytic center containing PLP covalently anchored to the enzyme via a conserved K208 residue and forming an internal aldimine as confirmed by the spectroscopic analysis (Fig. S1C and Fig. 2D,E). Importantly, loop L23–60 from a complementary subunit helps in defining the entrance into the catalytic cavity, and orienting PLP within the cleft. Specifically, residue R58 from the second subunit interacts with the phosphate group of the cofactor.
Finally, the C-terminal domain is organized into a five-stranded antiparallel β-sheet (exhibiting the following topology: ↑β8↓β9↑β12↓β11↓β10), decorated with five α-helices (α10–14) on one side which protect it from the solvent.
Similarly to all previously characterized CGLs, PaCGL self-assembles into a homotetramer that can be described as a dimer of dimers that include subunits A–B and C–D, respectively, in which the two subunits of each dimer are related by a 2-fold axis, and the dimers themselves maintain a 2-fold symmetry (Fig. 4D). The four subunits of the tetramer exhibit high similarity (average rmsd = 0.140 Å), differing only in the conformation of two long loops (L23–60 and L347–370), which appear partially disordered or present a slightly different conformation in some of the monomers. Notably, loop L347–370 from subunit A (Fig. 4D) adopts an extended conformation in PaCGL, diverging from the typical two-turn helix (α13) observed in the predicted AF2 model, and in the equivalent regions of CGLs from other species (Fig. S3A). On the other hand, loop L23–60 from the complementary B subunit of the A-B dimer was also nicely defined. This contrasts with the poor electron density observed for the region 46–57 in the other three subunits and similarly for the segment 350–366 in subunits B, C and D.
The extended conformation of the loop L347–370 directly impacts accessibility of the large cleft (chamber-2) located behind the PLP-containing catalytic site (chamber-1) (Fig. 5). Notably, the loop L347–370 does not display significant sequence conservation among various CGL homologs. Within helix α13, merely three amino acid residues remain conserved—specifically, proline, arginine, and glycine (P357, R361, G365 in PaCGL)—found within the loop´s second segment (Fig. S3). Strikingly, the AF2-predicted PaCGL model maintains the two-turns helicity fold present in other CGL enzymes (Fig. S3) suggesting the probability of two different (and stable) conformations for this segment. Differential conformation of this loop may modulate the accessibility of chamber-2, known to host inhibitors of Staphylococcus aureus CGL (SaCGL)4 and dictate the specificity of such inhibitors to PaCGL versus other species. Interestingly, the available crystal structures of various CGLs suggest a potential interdependence in the mobility of segments L23–60 and L347–370. Specifically, both loops appear either ordered or disordered simultaneously in the PaCGL crystals (Fig. S4), thereby influencing the accessibility of both the catalytic cavity (loop L23–60) and the chamber-2 (loop 347–370). To decipher the differential conformation of these loops, we compared the flexibility of each subunit within PaCGL tetramer with the equivalent regions in all available CGL homologs. The analysis, performed with CABSflex 2.027, compared the fluctuation plots for the different protein regions and confirmed a significantly higher mobility of the L23–60 and L347–370 regions in comparison with other protein segments. Moreover, these loops also showed the highest B-thermal parameters in the crystal structures of various CGLs (Fig. S5). The heightened mobility and potentially cooperative role of a third loop comprising residues 290–297 in PaCGL may potentially be involved in a novel regulatory mechanism modulating access to the chamber-2.
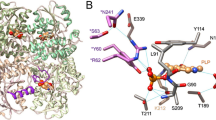
Main cavities and active site of PaCGL. (A) Main structural elements configuring the active site. Asterisks indicate elements from a complementary subunit. PLP is in sticks. The two main cavities, chamber-1 (active site) and chamber-2 (known to host inhibitors of SaCGL), are shown as red surface areas (Cavity detection cutoff = 4 solvent radii; Cavity detection radii = 7 Angstrom). (B) Stick representation of main amino acid residues within the PLP-binding cavity. Residues are colored according to the protein domain representation shown in Fig. 4. Polar interactions are represented in dashed lines. PLP is depicted in pink and red spheres correspond to water molecules. The Fo-Fc Polder omit electron density map (colored in light grey and contoured at 3σ) showing positive electron density around PLP is shown.
The active site of PaCGL (Figs. 4, 5) is located within a deep cavity of each monomer situated at the dimerization interface between subunits A–B (or equivalently, C–D) (Fig. 4), whose entrance is defined by four long loops. The first three (L205–219, L329–344, and L347–370) belong to the monomer containing the PLP cofactor, while the fourth (L23–60 and, more specifically, residues 40–60) belongs to the complementary monomer of the dimer. The cavity is completed with the N-terminal ends of α-helices containing residues 85–95 (α3) and 110–126 (α4) (Fig. 4). Helix α3 dipole moment helps orienting the phosphate moiety of PLP, while the side chain of the residue Y110 located on helix α4 packs parallel to the pyridine ring of the cofactor. On the other hand, loop L205–219 provides residue K208, a conserved lysine forming an internal aldimine with PLP cofactor, thus also determining the final orientation of PLP within the cavity. As the PaCGL tetramer assembly shows 222 symmetry, this particular arrangement of secondary elements is reproduced for each of the four catalytic cavities present in the CGL tetramer. Interestingly, loops L23–60 and L347–360 exhibit the highest mobility in all available CGL crystal structures to date (Fig. S4), reflecting a more pronounced fluctuation of the polypeptide chain (Fig. S5). These features suggest a potential regulatory mechanism as the conformation of these loops may limit accessibility of both the catalytic cavity and the chamber-2.
The main PLP-interacting residues, which are well-conserved across CGLs, are represented by Y110, R372, K208, S337, and S205 in PaCGL (Fig. 5). Residue K208 is covalently bound to PLP through its ε-amino group forming a Schiff-bond at the C4A position of PLP. The orientation of PLP is fixed by H-bond interactions between its phosphate group and the main chain nitrogen of residues G86 and L87. The hydroxyl group of S205 and T207 also stabilizes the PLP phosphate moiety. The complementary subunit interacts with O2P and O3P of PLP via the guanidine group of R58. Most CGLs also show interactions between the PLP and the conserved tyrosine of the neighboring subunit. In PaCGL, residue Y56 does not interact with the phosphate moiety of PLP, but rather establishes an H-bond with the N-terminal residue Q45. This contact is possible thanks to the flexibility provided by the residue G57. Interestingly, residue G57 of PaCGL is usually substituted by a conserved serine in other CGLs. Finally, the pyridoxal ring of PLP is stacked with the phenol ring of Y110.
Chamber-2 of PaCGL as potential drug binding site
The chamber-2 deserves a detailed analysis as it has been found to host pharmacological inhibitors in SaCGL4. Chamber-2 can be reached only via a narrow channel with limited accessibility determined by residues from the long loop connecting strands β11–β12 containing helix α13 (loop L347–370 in PaCGL), and by a either tyrosine (Y103 in SaCGL; Y102 in BcCGL), phenylalanine (F114 in PaCGL; F101 in LpCGL; Fig. 6, Fig. S3) or, alternatively, asparagine residue (N137 in TgCGL; N118 in HsCGL). The crystal structures of SaCGL complexed with novel pharmacological inhibitors NL1, NL2, and NL3 revealed the significance of the residue Y103 in SaCGL catalysis and its role in stabilizing the inhibitors within the cavity through a π-stacking interaction4. The Y103A mutation of SaCGL, or even the “humanized” Y103N variant, abolished the H2S-producing activity of SaCGL and disrupted the interaction with the inhibitors NL1 and NL24.
Chamber-1 and chamber-2 in PaCGL and SaCGL. (A) (Top) Surface representation of the PaCGL monomer. Chamber-1 and chamber-2 are colored in blue and magenta, respectively. (Bottom) Main amino acid residues within the cavities. PLP is in sticks. (B) (Top) Surface representation of the SaCGL monomer. Chamber-1- and chamber-2 are colored in blue and magenta, respectively. (Bottom) Main amino acid residues within the cavities. PLP is in sticks. NL1 inhibitor is in blue. The atom coordinates used to make the figure were obtained from PDB IDs 7BA4 (PaCGL) and 7MCT (SaCGL + NL1).
To assess the role of the equivalent chamber-2 residue F114 in PaCGL, we replaced it with alanine or asparagine and compared the resulting variants with the wild type. The two mutations did not impair the overall structural properties as well as the tetrameric oligomerization of the purified mutant enzymes (Fig. S6A–C). Strikingly, compared to SaCGL, both the F114A and the “humanized” F114N mutations resulted in only a 2–3 fold reduction in enzyme’s catalytic efficiency for both the canonical and H2S-producing activities of PaCGL (Table 2 and Fig. S6D).
Two additional amino acids situated at the base of the chamber-2 have been suggested to define the connecting gate between chamber-2 and the catalytic site (chamber-1). The first is a conserved histidine (H339 in SaCGL; H356 in HsCGL; H338 in BcCGL; H337 in LpCGL; and H376 in TgCGL) (Fig. 6). PaCGL also contains an equivalent histidine (H353 in PaCGL), but its spatial location differs from that found in other CGLs due to the more extended conformation of the loop L347–370, and the topological position of H339 in SaCGL is occupied by the M351 in PaCGL structure (Fig. 6A). The second residue connecting chamber-1 and chamber-2 is a conserved tyrosine that packs against the pyridine ring of PLP and helps to orient the cofactor (Y99 in SaCGL; Y114 in HsCGL; Y98 in BcCGL; Y97 in LpCGL; Y133 in TgCGL; and Y110 in PaCGL, respectively). The volume and access of chamber-2 differ significantly in SaCGL and PaCGL (Fig. 6B). In apo-HsCGL (no PLP cofactor present, PDB ID 3ELP), this tyrosine (Y114) appears displaced towards the bottom of the chamber-2 cavity, due to a partial unwinding of the last turn of helix α4. Upon binding of PLP (holo-HsCGL), the last turn of this same helix recovers its helicity and reorients the tyrosine towards the interior of the catalytic chamber-1. This conformational change is thought to function as an access gate from chamber-2 to the catalytic site.
Discussion
We conducted a biochemical and structural characterization of the CGL enzyme from P. aeruginosa. Our results clearly show an enzymatic competence of PaCGL to generate H2S using alternative substrates in addition to the canonical hydrolysis of l-Cth. Detailed structural comparison of PaCGL with CGL enzymes from other species revealed distinctive structural features within the primary cavities of the enzyme, which may modulate the access of substrates and/or inhibitors.
The canonical reaction of CGL within the transsulfuration pathway involves elimination of l-Cth at the γ-carbon. However, CGLs are prone to β-elimination of l-Cth as a side reaction. Our kinetic analysis demonstrated that PaCGL can catalyze both the α,γ- and α,β-cleavage of l-Cth to yield l-Cys and l-Hcys, respectively. Absorbance in the region of 440–480 nm in the UV–Vis and CD spectra in the presence of l-Cth provided support for the two elimination reactions. Spectral features of the PaCGL-substrate complex in the long-wavelength region are similar to those of the Citrobacter freundii methionine γ-lyase complex with methionine28,29. Two overlapping absorption bands with maxima at ~ 460 and ~ 485 nm were also observed in the yeast CGL30. The absorption at 480 nm likely corresponds to the formation of α-aminocrotonate, an essential intermediate of the γ-elimination reaction30, while absorption at around 460 nm is probably due to α-aminoacrylate formation, which has been observed under steady-state conditions for β-elimination reactions of different PLP-dependent enzymes30,31,32.
PaCGL exhibited a notably high catalytic efficiency for the hydrolysis of l-Cth (17 mM−1 s−1), surpassing the activities of CGLs from other organisms (LbCGL, 1.1 mM−1 s−1; TgCGL, 2.2 mM−1 s−1; BcCGL, 3.2 mM−1 s−1; ScCGL, 2.1 mM−1 s−1; HsCGL, 8.2 mM−1 s−1)17,19,22,24,33,34.
PaCGL also produces H2S using l-Cys or/and l-Hcys as alternative substrates. This aligns with the previous reports showing that inactivation of the Cse gene, encoding for the PaCGL, leads to a reduction of H2S production in P. aeruginosa4. We investigated various reactions catalyzed by PaCGL that lead to H2S biogenesis and the uncommon thioether-bond-containing amino acids l-Lanthionine and l-Homolanthionine. We found that PaCGL efficiently produced H2S via different mechanisms such as (i) β-elimination of l-Cys into H2S and l-Ser, (ii) β-replacement of 2 mol of l-Cys forming H2S and l-Lanthionine, (iii) γ -elimination of l-Hcys into H2S and l-Homoserine, (iv) γ-replacement of 2 mol of l-Hcys forming H2S and l-Homolanthionine, and (v) the replacement of l-Hcys and l-Cys producing H2S and l-Cth. Under substrate saturating conditions, the catalytic efficiency (kcat/Km) for the H2S elimination from l-Cys and l-Hcys is approximately 25- and 68-fold lower, respectively, than for the canonical hydrolysis of l-Cth (Table 1). However, the occurrence and regulation of these alternative reactions in the cell remain unknown. The bacterial intracellular concentrations of l-Cys are tightly regulated. Notably, P. aeruginosa displays a high redundancy in l-Cys production. In addition to the CGL and CBS enzymes in the RTP, this pathogen possesses genes encoding the enzymes for the de novo l-Cys synthesis pathway, i.e., serine acetyltransferase (SAT) catalyzing the condensation of l-Ser and the acetyl group of acetyl-CoA to form O-acetylserine (OAS), and the cysteine synthase (CS), which catalyzes the nucleophilic attack of sulfide (H2S) on OAS to form l-Cys and releasing acetate. l-Cys is also known to participate in the allosteric inhibition of SAT, leading to the production of OAS. OAS, in turn, is converted into N-acetylserine, an auto-inducer of the transcription regulator (the CysB protein), which acts as a sensor and regulator of the intracellular content of l-Cys and sulfur35. Further studies are required to determine whether PaCGL serves as the primary checkpoint in the RTP in P. aeruginosa and its involvement in l-Cys production.
Shatalin et al.4 solved crystal structures of SaCGL complexed with three novel inhibitors NL1, NL2, and NL3 demonstrating high specificity towards bacterial CGL and no impact on human (host) enzyme. Our crystal structure of PaCGL enabled us to compare essential regions of this enzyme (chamber-1 and chamber-2) with the corresponding regions in other bacterial counterparts (including S. aureus) and the human enzyme. The analysis revealed distinct structural characteristics that set PaCGL apart, potentially paving the way for future drug development targeting this important metabolic enzyme. While chamber-1, corresponding to the catalytic site, is highly conserved among CGLs and PLP-dependent enzymes in general, chamber-2 has unique physical–chemical properties and its distinct conformation sets PaCGL apart from the other CGLs (Fig. 6). An intriguing difference in the chamber-2 of PaCGL is the presence of the residue F114 (Fig. 6A), which occupies equivalent position of the conserved residue Y103 in SaCGL (Fig. 6B). Strikingly, the F114A mutation in PaCGL did not result in loss of enzyme activity, as it occurred for the equivalent Y103A mutation in SaCGL, supporting the notion that this residue not only determines the general characteristics of the entrance channel to chamber-2 (volume, size, steric hindrance), but modulates the overall volume of the chamber and, consequently, the type of molecule that can be accommodated within the chamber-2. In addition, the conformation of a long loop containing helix α13 (L347–370 in PaCGL) is important for accessibility of the chamber-2 (Fig. 5). This loop determines the width of the chamber-2 entrance and affects the void volume of the entire cleft (Fig. S7). Interestingly, our crystal structure of PaCGL revealed that the helicity of this loop is partially lost, resulting in a more extended peptide segment that allows for a wider access into chamber-2 (a comparison of the internal cavity volume for chamber-2 and the fold of the equivalent regions to loop L347–370 in CGLs from different organisms is shown in Fig. S7). However, despite the cavity opening being bigger, the reorientation and shape of the loop L347–370 in PaCGL reconfigured the internal contour and, consequently, made the volume of the cavity smaller compared to what was observed, for example, in SaCGL (loop L332–356) or the human enzyme (loop L349–373) (Fig. 6 and Fig. S7). The comparison of the crystal structures of PaCGL and SaCGL complexed with NL1, NL2, and NL3 inhibitors suggests that the distinct arrangement of PaCGL observed in this region would not hinder the binding of these molecules to PaCGL, requiring only minor shifts of side chain to accommodate them inside.
The PaCGL model predicted by AF2 (Fig. S7) exhibited a helical conformation of the loop L347–370 similar to that of other CGLs but different from the conformation found in our PaCGL crystals. This suggests that the opening and closing of the chamber-2 cavity may differ from what was proposed based on the apo- and holo-states of the human enzyme. As demonstrated in Fig. 6, Fig. S7, and Movie S1, the increased helicity of this loop in the PaCGL AF2 model corresponds to a closed conformation of the chamber-2 with a restricted accessibility and volume. The H353 residue would occupy a position like that found in other CGLs. In contrast, the extended conformation observed in the crystals made the chamber-2 larger, thus likely more accessible for small molecules, such as NL1, NL2, and NL3 inhibitors. Interestingly, in this open state, the residue M351 occupies the equivalent position to the conserved histidine in other CGLs, suggesting its role in modulating the type of molecules that can be hosted inside the cavity. Future molecular dynamics studies could provide additional new insights into conformational arrangement of the chamber-2.
Overall, our findings revealed the fundamental structural traits of the P. aeruginosa CGL enzyme, which has gained significant attention as a potential pharmacological target due to its role in H2S biogenesis in this emerging and concerning pathogen.
Experimental section
Protein production
Gene sequence encoding for PaCGL (PAO1_PA0400) with a N-terminal 6x-His Tag was synthesized by Genscript, PCR amplified and cloned into a modified pET28a expression vector (Novagen). The F114A and F114N point mutations were introduced by site-directed mutagenesis using QuikChange II Kit (Agilent), using the primers in Table S1. All constructs were verified by DNA sequencing performed by Eurofins Genomics. The PaCGL constructs were transformed into E. coli Rosetta (DE3) expression host cells (Novagen). Cells were grown in Luria–Bertani medium at 37 °C to a turbidity of 0.6–0.8 at 600 nm. Expression was induced with 0.5 mM IPTG for 16 h at 24 °C. Cells were harvested and resuspended in 20 mM sodium phosphate pH 8.0, 150 mM NaCl, 0.1 mM DTT containing a protease inhibitor cocktail EDTA free. After sonication, the suspension was centrifuged at 30,000×g for 20 min at 4 °C. The supernatant was recovered and loaded on an Ni–NTA Sepharose column (GE-Healthcare) equilibrated with 20 mM sodium phosphate pH 8.0, 150 mM NaCl, 0.1 mM DTT and 10 mM imidazole. A linear gradient from 10 to 500 mM imidazole was then applied. Fractions enriched in PaCGL were pooled together, concentrated and buffer exchanged into 20 mM sodium phosphate pH 8.0, 150 mM NaCl, 0.1 mM DTT buffer using Vivaspin concentrators (Sartorius). Each purification yielded about 100 mg of pure protein per liter of bacterial culture. To facilitate crystallization, an additional construct for PaCGL wild-type with the C-terminal 6x-His tag was prepared using similar strategy as described above.
The theoretical extinction coefficient of monomeric PaCGL at 280 nm was 28,545 M−1 cm−1 (http://www.expasy.ch/tools/protparam.html). The PLP content of the enzyme was determined by releasing the coenzyme in 0.1 M NaOH and by using εM = 6600 M−1 cm−1 at 388 nm.
The oligomeric state of PaCGL variants was determined by gel filtration using a Sephacryl S-200 16/60 HR column in 20 mM sodium phosphate pH 8.0, 150 mM NaCl and 0.1 mM DTT. The calibration curve was generated following the protocols in36,37.
The apo-form of PaCGL was obtained by incubating the enzyme with phenylhydrazine hydrochloride following the protocol in38. The dissociation constant for PLP (Kd) was obtained by monitoring the change of intrinsic fluorescence (excitation was set at 295 nm) of 1 μM apo-protein at different concentrations of PLP (0.01–4 μM) in 20 mM sodium phosphate pH 8.0 at 25 °C on a FP8200 Jasco spectrofluorimeter24,39.
Spectroscopic measurements
Absorption spectra of 15 μM PaCGL were collected on a Jasco V-750 UV–visible spectrophotometer in 20 mM sodium phosphate pH 8.0 at 25 °C31. CD spectra were recorded on CD spectropolarimeter Jasco J-1500 equipped with a Peltier type thermostated cell holder, as previously described31,40. Briefly, far-UV (190–250 nm) spectra of 0.2 mg mL−1 PaCGL variants were collected in using a 0.1-cm path length quartz cuvette. Near UV–Vis (250–600 nm) spectra of 1 mg mL−1 PaCGL variants were recorded in 1-cm path length quartz cuvette at 25 °C. A minimum of three accumulations were made for each scan, averaged, and corrected for the blank solution of corresponding buffer. Thermal unfolding profiles were collected by recording ellipticity at 222 nm in a temperature range between 15 to 90 °C (scan rate 90 °C/h) using 0.1-cm path length quartz cuvettes and protein concentration of 0.2 mg mL−1. All CD measurements were recorded in 20 mM sodium phosphate pH 8.041.
Enzyme activity assays
The CGL activity in the l-Cth γ-elimination reaction was determined by a previously described 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) assay24,42.
Briefly, reactions (200 µL sample) were carried out at 37 °C in assay buffer (50 mM MOPS, 50 mM bicine, 50 mM proline pH 8.0, 20 μM PLP) containing 0.2 mM DTNB in the presence of different concentrations of l-Cth (0–8 mM). The reactions were initiated by the addition of PaCGL to 1 μM. Absorbance changes were monitored in continuous at 412 nm (Δε412 = 13,600 M−1 cm−1).
The activities in the H2S-generating alternative reactions were measured using the lead acetate assay as described elsewhere31,43,44,45. The enzyme (1–4 µM) was added to 0.4 mL of reaction mixture containing 50 mM HEPES pH 7.4, 20 μM PLP, 0.4 mM lead (II) acetate, and 0–50 mM l-Cys or 0–50 mM l-Hcys.
Pyruvate formation was measured by monitoring NADH oxidation (ε340 = 6200 M−1 cm−1) via LDH assay36. Data for H2S production from l-Cys by PaCGL were fitted following the kinetic models described previously19,45. Briefly, H2S production from l-Cys is the sum of two possible reactions, the β-elimination of l-Cys to generate l-Ser (reaction 3) or the condensation of two molecules of l-Cys to generate l-Lanthionine (reaction 4). Data for overall H2S production from L-Cys were fitted using Eq. (1) where vl-ser and vLanthionine are defined by Eq. (2) and Eq. (3), respectively 19,31,45.
where Km1 and Vmax1 are associated to the unimolecular reaction, Km2 and Vmax2 to substrate binding at the second site and the reaction velocity of the bimolecular reaction and n represents Hill coefficient.
Liquid chromatography mass spectrometry (LC–MS/MS)
A TSQ Fortis Triple Quadrupole mass spectrometer (Thermo Scientific) coupled to Ultimate 3000 HPLC system (Thermo Scientific) was used for this analysis. The products separation was performed on a Luna C18(2) column (150 × 4.6 mm, 3 µm particle size, Phenomenex) with gradient elution. The mobile phase was composed of formic acid (A, 0.1% formic acid in water) and acetonitrile (B, 0.1% formic acid in ACN). Chromatographic gradient elution was the following: constant flow of 0.4 mL min−1; 98% phase A at time 0, then decreased up to 5% A in 10 min and maintained at 5% A for 2 min and re-equilibrated for 5 min. The ESI source settings were ion spray voltage, +3500 V; ion transfer tube, 300 °C; sheath gas and aux gas, 50 and 10, respectively, vaporizer temperature 350 °C. Multiple reaction monitoring was optimized using nitrogen as collision gas (with pressure set at 1.5 mTorr). Two transitions for each substance were chosen for identification. Data acquisition and elaboration were performed by the Chromeleon (version 7.2, Thermo Fisher).
Protein crystallization
For crystallization, the enzymes were buffer exchanged into 50 mM HEPES, 150 mM NaCl, 0.1 mM DTT pH 8.0. Preliminary crystallization trials were carried out by the vapor-diffusion technique in a sitting drop format with 96-well MRC crystallization plates, following a previously described protocol22.
Drops consisted of 200 nL protein solution (20 mg mL−1) were mixed with 200 nL precipitant solution and incubated at 293 K. The successful condition was scaled-up in a hanging-drop format using 24-well VDX plates (Hampton Research) in a reservoir with drops consisting of 0.5 μL protein (protein concentration of 20 mg mL−1) with 0.5 μL precipitant solution. This reservoir was composed of 9% w/v PEG 4000 and 0.1 M sodium acetate pH 4.6 with a volume of 0.5 mL. The crystals were transferred to a crystallization buffer containing 9% (w/v) PEG 4000, 0.1 M sodium acetate pH 4.6, and 20% glycerol for a few seconds before being cryocooled in liquid nitrogen.
Structural determination by X-ray crystallography
All X-rays datasets were collected at Synchrotron beamlines XALOC (ALBA), I03/I24 (DIAMOND, UK) and ID29 of ESRF (Grenoble). Datasets were collected over a range of 0.1–0.25° and the distance to the detector was set to reach resolution data between 1.6–3.8 Å depending on the crystal, and according to the diffraction parameters previously determined by several test images. Several data set were collected but only one allowed the structural determination of PaCGL (Table S2). Diffraction data were processed using HKL200046 or XDS47 programs. The three-dimensional structure of PaCGL was determined by MR method with the Phaser-MR program48 from Phenix Suite49 using the coordinates of HsCGL holoenzyme (PDB ID 2NMP) as initial search model. The geometric quality of the models was assessed with MolProbity50 integrated in Phenix suite. Figures were done with Pymol (The PyMOL Molecular Graphics System, Version 2.2.3, Schrödinger, LLC)51 and UCSF Chimera52.
Deep learning structural comparison
Protein structure predictions were performed with AlphaFold 2.3.025 using an adapted version of the AF2 code (https://github.com/deepmind/alphafold).
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files. The nucleotide sequence of PaCGL (PAO1_PA0400) can be accessed in the Pseudomonas aeruginosa database (https://www.pseudomonas.com/feature/show?id=103537). Additionally, the crystal structure of PaCGL (PDB code: 7BA4) is accessible through the Protein Data Bank (https://www.rcsb.org/structure/7BA4).
References
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18(3), 318–327 (2018).
Sadikot, R. T., Blackwell, T. S., Christman, J. W. & Prince, A. S. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171(11), 1209–1223 (2005).
Shatalin, K., Shatalina, E., Mironov, A. & Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 334(6058), 986–990 (2011).
Shatalin, K. et al. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science 372(6547), 1169–1175 (2021).
Mironov, A. et al. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. 114(23), 6022–6027 (2017).
Nzungize, L. et al. Mycobacterium tuberculosis metC (Rv3340) derived hydrogen sulphide conferring bacteria stress survival. J. Drug Target. 27(9), 1004–1016 (2019).
Rahman, M. A. et al. The role of host-generated H2S in microbial pathogenesis: New perspectives on tuberculosis. Front. Cell. Infect. Microbiol. 10, 586923 (2020).
Walsh, B. J. C. & Giedroc, D. P. H2S and reactive sulfur signaling at the host-bacterial pathogen interface. J. Biol. Chem. 295(38), 13150–13168 (2020).
Chang, W. et al. Inhibition of fungal pathogenicity by targeting the H2S-synthesizing enzyme cystathionine β-synthase. Sci. Adv. 8(50), eadd5366 (2022).
Croppi, G. et al. Discovery of an inhibitor for bacterial 3-mercaptopyruvate sulfurtransferase that synergistically controls bacterial survival. Cell Chem. Biol. 27(12), 1483-1499.e9 (2020).
Seregina, T. A., Lobanov, K. V., Shakulov, R. S. & Mironov, A. S. Enhancement of the bactericidal effect of antibiotics by inhibition of enzymes involved in production of hydrogen sulfide in bacteria. Mol. Biol. 56(5), 638–648 (2022).
Fang, D., Wang, Z. & Liu, Y. Cystathionine γ-lyase: The Achilles heel of bacterial defense systems. Int. J. Antimicrob. Agents 62(1), 106845 (2023).
Weikum, J. et al. Sulfide protects Staphylococcus aureus from aminoglycoside antibiotics but cannot be regarded as a general defense mechanism against antibiotics. Antimicrob. Agents Chemother. 62(10), e00602-18 (2018).
Caruso, L. et al. Hydrogen sulfide production does not affect antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 6, e0007524 (2024).
Muyzer, G. & Stams, A. J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6(6), 441–454 (2008).
González-Recio, I., Fernández-Rodríguez, C., Simón, J., Goikoetxea-Usandizaga, N., Martínez-Chantar, M. L., Astegno, A., Majtan, T. & Martinez-Cruz, L. A. current structural knowledge on cystathionine β-synthase, a pivotal enzyme in the transsulfuration pathway. in eLS, 453–468 (2020).
Matoba, Y. et al. Catalytic specificity of the Lactobacillus plantarum cystathionine γ-lyase presumed by the crystallographic analysis. Sci. Rep. 10(1), 14886 (2020).
Steegborn, C. et al. Kinetics and inhibition of recombinant human cystathionine gamma-lyase. Toward the rational control of transsulfuration. J. Biol. Chem. 274(18), 12675–12684 (1999).
Chiku, T. et al. H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 284(17), 11601–11612 (2009).
Metzler, C. M., Harris, A. G. & Metzler, D. E. Spectroscopic studies of quinonoid species from pyridoxal 5′-phosphate. Biochemistry 27(13), 4923–4933 (1988).
Woehl, E. U., Tai, C. H., Dunn, M. F. & Cook, P. F. Formation of the alpha-aminoacrylate immediate limits the overall reaction catalyzed by O-acetylserine sulfhydrylase. Biochemistry 35(15), 4776–4783 (1996).
Fernández-Rodríguez, C. et al. Structural basis of the inhibition of cystathionine γ-lyase from Toxoplasma gondii by propargylglycine and cysteine. Protein Sci. 32(4), e4619 (2023).
Yamagata, S., Isaji, M., Yamane, T. & Iwama, T. Substrate inhibition of l-cysteine alpha, beta-elimination reaction catalyzed by l-cystathionine gamma-lyase of Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 66(12), 2706–2709 (2002).
Maresi, E. et al. Functional characterization and structure-guided mutational analysis of the transsulfuration enzyme cystathionine γ-lyase from Toxoplasma gondii. Int. J. Mol. Sci. 19(7), 2111 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596(7873), 583–589 (2021).
Liang, J., Han, Q., Tan, Y., Ding, H. & Li, J. Current advances on structure-function relationships of pyridoxal 5′-phosphate-dependent enzymes. Front. Mol. Biosci. 6, 4 (2019).
Kuriata, A. et al. CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 46(W1), W338–W343. https://doi.org/10.1093/nar/gky356 (2018).
Morozova, E. A. et al. Kinetic and spectral parameters of interaction of Citrobacter freundii methionine γ-lyase with amino acids. Biochemistry (Moscow) 75(10), 1272–1280 (2010).
Anufrieva, N. V. et al. The role of active site tyrosine 58 in Citrobacter freundii methionine γ-lyase. Biochim. Biophys. Acta BBA Proteins Proteomics 1854(9), 1220–1228 (2015).
Yamagata, S., Yasugahira, T., Okuda, Y. & Iwama, T. Conversion of the aminocrotonate intermediate limits the rate of γ-elimination reaction catalyzed by l-cystathionine γ-lyase of the yeast Saccharomyces cerevisiae. J. Biochem. 134(4), 607–613 (2003).
Conter, C. et al. Cystathionine β-synthase is involved in cysteine biosynthesis and H(2)S generation in Toxoplasma gondii. Sci. Rep. 10(1), 14657 (2020).
Jhee, K. H., Niks, D., McPhie, P., Dunn, M. F. & Miles, E. W. The reaction of yeast cystathionine beta-synthase is rate-limited by the conversion of aminoacrylate to cystathionine. Biochemistry 40(36), 10873–10880 (2001).
Hopwood, E. M., Ahmed, D. & Aitken, S. M. A role for glutamate-333 of Saccharomyces cerevisiae cystathionine gamma-lyase as a determinant of specificity. Biochim. Biophys. Acta 1844(2), 465–472 (2014).
Sagong, H.-Y., Kim, B., Joo, S. & Kim, K.-J. Structural and functional characterization of cystathionine γ-lyase from Bacillus cereus ATCC 14579. J. Agric. Food Chem. 68(51), 15267–15274 (2020).
Kredich, N. M. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6(19), 2747–2753 (1992).
Astegno, A., Giorgetti, A., Allegrini, A., Cellini, B. & Dominici, P. Characterization of C-S Lyase from C. diphtheriae: A possible target for new antimicrobial drugs. Biomed Res. Int. 2013, 701536 (2013).
Astegno, A., Capitani, G. & Dominici, P. Functional roles of the hexamer organization of plant glutamate decarboxylase. Biochim. Biophys. Acta 1854(9), 1229–1237 (2015).
Allegrini, A., Astegno, A., La Verde, V. & Dominici, P. Characterization of C-S lyase from Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 and its potential role in food flavour applications. J. Biochem. 161(4), 349–360 (2017).
Astegno, A. et al. Unique substrate specificity of ornithine aminotransferase from Toxoplasma gondii. Biochem. J. 474(6), 939–955 (2017).
Bombardi, L., Pedretti, M., Conter, C., Dominici, P. & Astegno, A. Distinct calcium binding and structural properties of two centrin isoforms from Toxoplasma gondii. Biomolecules 10(8), 1142 (2020).
Trande, M. et al. Cation and peptide binding properties of CML7, a calmodulin-like protein from Arabidopsis thaliana. J. Inorg. Biochem. 199, 110796 (2019).
Aitken, S. M. & Kirsch, J. F. Kinetics of the yeast cystathionine beta-synthase forward and reverse reactions: Continuous assays and the equilibrium constant for the reaction. Biochemistry 42(2), 571–578 (2003).
Banerjee, R. et al. Assay methods for H2S biogenesis and catabolism enzymes. Methods Enzymol. 554, 189–200 (2015).
Conter, C. et al. Insights into domain organization and regulatory mechanism of cystathionine beta-synthase from Toxoplasma gondii. Int. J. Mol. Sci. 23(15), 8169 (2022).
Singh, S., Padovani, D., Leslie, R. A., Chiku, T. & Banerjee, R. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 284(33), 22457–22466 (2009).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62(Pt 8), 859–866 (2006).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66(2), 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40(Pt 4), 658–674 (2007).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75(Pt 10), 861–877 (2019).
Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66(Pt 1), 12–21 (2010).
DeLano, W. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25(13), 1605–1612 (2004).
Acknowledgements
This research was supported by the MUR-PRIN 2022 Grant No. 20224BYR59 to A.A., by the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4—Call for tender No. 341 of 03.15.2022 of Italian Ministry of University and Research funded by the European Union—NextGenerationEU to A.d.M., by Spanish Ministry of Economy and Competitiveness Grant BFU2016-77408-R and by Spanish Ministerio de Ciencia e Innovación (MICINN), Grants No PID2019-109055RB-I00 and PID2022-141748OB-I00, to L.A.M.-C. We also thank MINECO for the Severo Ochoa Excellence Accreditation (CEX2021-001136-S). T.M. acknowledges the support from University of Fribourg Research Pool grant (22-15). We thank the Centro Piattaforme Tecnologiche of the University of Verona for providing access to the spectroscopic and mass spectrometry platforms.
Author information
Authors and Affiliations
Contributions
M.P., C.F.-R, C.C. and I.O. conceived the study, conducted the experiments, analysed the data, and edited the manuscript. F.F., A.d.M., P.D., M.L.M-C. and M. PET. contributed to the discussion of results and edited the manuscript. M.L.M.-C., T.M. supported funding acquisition and edited the manuscript. A.A. and L.A.M.-C. conceived the study, provided financial support, analysed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pedretti, M., Fernández-Rodríguez, C., Conter, C. et al. Catalytic specificity and crystal structure of cystathionine γ-lyase from Pseudomonas aeruginosa. Sci Rep 14, 9364 (2024). https://doi.org/10.1038/s41598-024-57625-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-57625-7
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.