Abstract
African yam bean (AYB) (Sphenostylis stenocarpa (Hochst ex. A. Rich.) harms) an underutilized legume that produces nutritionally healthy seeds and tubers in some variety. The low yield of the crop is attributed to production constraints such as attacks by pest and disease-causing organisms such as fungi, bacteria and viruses. In this study, one hundred AYB accessions were evaluated for resistance to viral infection. The AYB accessions were planted using a randomized complete block design on the experimental field at the International Institute of Tropical Agriculture (IITA) Ibadan, Nigeria. Viral disease severity was assessed at 10, 12, 14, 16 and 18 weeks after planting (WAP) based on disease symptoms using disease severity index on visual scale of 1–5. Antigen–coated plate enzyme linked immunosorbent assay (ELISA) and reverse transcription polymerase chain reaction were used to index diseased leaf samples collected from the field. Result from five virus species (Cowpea mild mottle virus, Cowpea mottle virus, Southern bean mosaic virus, Cowpea mosaic virus and Bean common mosaic virus) were detected in few accessions while mixed infections were observed in some accessions. TSs-552, TSs-577, TSs-580, TSs-560 and TSs-600 were devoid of viruses and could be resistant. There were no significant differences at p < 0.05 in the mean disease incidence (DI) of viral diseases. However, at 18 weeks after planting, TSs-604 had the highest (100%) mean DI while TSs-584 had the lowest (13.33%) mean DI. Cluster analysis based on the AUDPC produced 6 main clusters, the clusters revealed grouping patterns in which AYB lines with similar resistance ratings were shown to form unique clusters. The information generated from this study will contribute to the development of strategies in the management of virus diseases infecting AYB.
Similar content being viewed by others
Introduction
African yam bean (Sphenostylis stenocarpa (Hochst ex. A. Rich.) harms) is a nutritionally important but neglected food crop with several benefits. The crop has the ability to produce bean seed in a pod with varying seed patterns and colours1. In addition to the seeds, farmers can also harvest tubers, which resemble sweet potato. The tubers mature in 5 to 8 months2. The edible root tubers are rich in nutrients suitable for human consumption3. One of the limiting factors to the production of African yam bean is it low grain yield when compared with other legumes4. Hence, the low yield of the crop is attributed to production constraints such as unavailability of improved seeds, poor farm practices, attack by pest and disease-causing organisms such as fungi, bacteria and which viruses pose a major concern to African yam bean production. Viral diseases are among the most important pathogens in agriculture and have the ability to cause great economic losses to farmers by affecting the yield quality of the crop5,6. Viruses pose serious risks and may restrict the international movement of improved or selected germplasm due to quarantine restrictions7. Viral infected plant produce little or no flower buds and pod8. Several viruses have been reported worldwide, of which two viruses have been identified serologically infecting AYB9. This includes cowpea mild mottle virus (CPMMV) and black eye cowpea mosaic virus (BCMV). Symptoms of viral diseases in AYB include mosaic, distortion, yellow chlorosis and stunting of the plant. Effective management of these viral diseases are quite important to improve yield of AYB. Conventional methods used by farmers to control viruses including broad-spectrum insecticides for the control of vectors that transmit the viruses are inadequate and not cost-effective to the farmers and not environmentally friendly. The use of host plant resistance is therefore considered the most economical and environmentally friendly approach in the management of viral diseases. This study was conducted to evaluate one hundred AYB accessions and to identify the associated viruses using serological and RT-PCR techniques. Detection of AYB virus will be important in developing measures to control the virus infecting AYB.
Results
Disease symptoms observed
Virus and virus-like disease symptoms were observed on 100 AYB accessions on the field. The symptoms included different symptoms such as mosaic, puckering, stunting, leave curl, chlorosis and mottling (Fig. 1). Over 88.33% of the accessions showed mosaic followed by mottling (40.7%), Leaf curl (39.7%), puckering (20.3%), stunting (17.3%) whereas chlorosis was the least (9%) (Fig. 2).
Incidence of viral diseases
Mean incidence of viral diseases on the 100 accessions of African yam bean is presented in Table 1. Generally, for all 100 African yam bean accessions, the incidence of viral diseases increased from 10 to 18 WAP, with overall mean incidences increasing from 27.01 to 60.39%. ANOVA showed no significant differences in the mean incidence of viral diseases during the 2019/2020 planting season. At the final observation (18 WAP), TSs-601 had the highest mean incidence (93.33%), while TSs-584 had the lowest mean incidence of 13.33%.
The severity of viral diseases and area under disease progress curve
The results of cluster analysis performed based on the severity of the viral symptoms and area under disease progressive curves (AUDPC) are presented in Fig. 3 the 100 accessions were grouped into six major clusters. Cluster 1 has 8 accessions which are highly susceptible to the observed viral symptoms based on AUDPC. Cluster 5 comprises of 32 accessions that were all moderately resistant to viral symptoms.
Viral detection by antigen-coated plate enzyme-linked immunosorbent assay (ACP-ELISA) and reverse transcriptase polymerase chain reaction (RT-PCR)
Five virus species namely CPMMV, CPMoV, SBMV, CMV, and BCMV were detected in the African yam bean accessions using ACP-ELISA. RT-PCR detected BCMV in several accessions and CAbMV was not detected in the accession. Seventy-five sample out of the eighty symptomatic samples collected was infected with at least one of the five viruses (Table 2). It was observed that some viruses were associated with single or multiple infections in the plant samples. The AYB samples had a high prevalence of single virus infection compared with multiple virus infections. In single virus-infected leaf samples, BCMV was the most prevalent, infecting 73.75% of the samples tested, Co-infection by BCMV + CPMMV was observed in twelve African yam bean accessions. BCMV, CPMMV, and SBMV were observed in two AYB accession. BCMV + CPMMV + SBMV + CMV were observed in one accession of AYB; five accessions of AYB TSs-552, TSs-577, TSs-580, TSs560, and TSs600 were not positive for any of the four antibodies used in this study (Fig. 4).
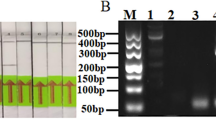
Agarose gel electrophoresis (2% agarose) of resolved amplified RT-PCR products of BCMV polyprotein gene (469 bp). Stained with ethidium bromide. Lane M: Marker 100 bp DNA; Lanes 1–18 test samples; Lanes 19–37 test samples; Lane 38–55 test samples; lane 56–72 test sample; lane 73–83 test samples, H healthy control, and D disease control.
Discussion
The success of disease resistance breeding relies on precise identification of pathogen and accurate screening of germplasm lines for a particular disease-causing pathogen. It is preferable to adopt disease management strategies including breeding resistant plants, limiting the movement of plant material, and searching for new variants to lessen the damage that plant-pathogenic viruses can cause10,11. This study was conducted to evaluate one hundred AYB accessions and to identify the associated viruses. The AYB accessions exhibited various symptoms, including leaf mosaic, mottling, chlorotic, leaf curl, puckering and chlorosis. The presence of these symptoms suggested that AYB was infected with viruses on the field. Several types of symptoms have been reported on virus infecting leguminous plant12,13,14. The symptoms observed in this study on AYB accessions were in consistent with the symptom types that has been previously reported on AYB9. The various African yam bean symptoms observed in the field may be due to factors such as the genotype and type of virus, the time of infection of the virus pathogen, mixed infections and environment. Four viruses infecting African yam bean was detected using ACP-ELISA namely CMV, SBMV, CMoV and CPMMV. These viruses can cause great damage on plant. RT-PCR was used for detecting the presence of BCMV. Bean common mosaic virus (BCMV) is one of the most prevalent and harmful viruses, affecting both cultivated and wild range of leguminous plant15. The viruses identified in this study are among the virus listed by Hughes et al. to be occurring in Nigeria16. Fifty-nine accessions out of the eighty symptomatic accessions were infected with BCMV. This suggest that BCMV may be the most common virus infecting African yam bean in the study area. One of the major causes of the reported low yield in AYB could be attributed to BCMV. The study also revealed mixtures of infection within the African yam bean accessions. Several reports have shown that multiple virus infections are usually associated with higher disease severity and yield reduction17,18. Virus-symptomatic plants not testing positive to any virus for which diagnostics were used in this study may be due to their low concentrations in the leaf samples19.
Conclusions
This study demonstrated that leaf mosaic and mottling were the most common symptoms observed in AYB followed by chlorotic spots and the least was stunting. ACP-ELISA and RT-PCR tests detected five viruses in the accessions studied. In contrast, no virus was detected in accessions TSs-552, TSs-577, TSs580, TSs-560 and TSs-600 by ACP-ELISA and RT-PCR which may imply diseases resistance in these five African yam bean accessions for viruses tested. These accessions can be further screened against other African yam bean viruses to confirm their status. Confirmation of the resistance of these accessions may serve as a source of resistant genes for breeding work and planting material for farmers.
Materials and methods
All methods used in this study were carried out in accordance with relevant guidelines and regulations.
The research was conducted at the experimental field of the International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria. One hundred accessions of African yam bean were obtained from the Genetic Resources Center of IITA Ibadan, Nigeria (Fig. 5). Seeds were planted on 5 m ridges, spaced 1 m apart. Each accession was planted on two rows at 1 m intra-row spacing. Initially, two seeds were planted per hill and later thinned to one plant per hill to give 10 plants per accession in each plot. The experiment was prepared in three replicates. Disease assessment was done 14 days interval starting from the fourth weeks after sowing (WAS) until senescence. (Table 3). Data on mean severity scores were used to calculate Area Under Disease Progress Curve for each of the African yam bean lines using the formula of Campbell and Madden20.
where n was the total number of assessments, ti was the time of the ith assessment in days from the first assessment date, xi was percentage of disease severity at ith assessment.
Serological detection
Leaf samples collected from symptomatic and asymptomatic plants were tested with antigen-coated-plate ELISA (ACP-ELISA) using six polyclonal antibodies specific for legumes; Cowpea mild mottle virus (CPMMV), Cowpea mottle virus (CMV), Southern bean mosaic virus (SBMV), Cowpea mottle virus (CPMoV), Bean common mosaic virus (BCMV) and Cowpea Aphid-borne mosaic virus (CABMV). The ACP ELISA was carried out as described by Kumar21. African yam bean leaf samples were ground at a ratio of 0.1 g ml–1 (1:10 w/v) with a mortar and pestle in coating buffer (Na2CO3 1.59 g, NaHCO3 2.93 g and sodium diethyldithiocarbamate 10 g in 1 L of distilled water with pH 9.6). Hundred micro liters (100 ul) of p- nitrophenyl phosphate substrate was added to each well and incubated at room temperature for 1 h, and overnight at 4 °C to allow color development. Optical density values were read at 405 nm using a Bio-Rad microplate reader (ELx 800, Universal Microplate Reader). The result was considered positive when the value is greater or equal to twice the absorbance value of healthy control.
Detection by reverse transcription polymerase chain reaction (RT-PCR)
Samples confirming virus positive were amplified by modified reverse transcriptase-polymerase chain reaction (RT-PCR) protocol using only two coat protein specific primers (Cowpea aphid-borne mosaic virus (CABMV) − 525bp and Bean common mosaic virus (BCMV)—469bp) RT-PCR was set up for BICMV and CABMV-positive samples in ACP-ELISA, using primers designed to coat protein as follows BCMVF3X 5′-ATGTGGTACAATGCTGTGAAG, BCMV B3X TTTCAGTATTCTCGCTGGTTG. CABMV coat protein gene, CABMV F3X 5′- GTACTCCAGTCTGATGGAAAGG, CABMV B3X GTCCGAGAAGTGGTGCATAA. Total nucleic acid was extracted from African yam bean leaf samples using modified Cetyltrimethyl Ammonium Bromide (CTAB) method22. The extracted RNA was used as template in RT-PCR to amplify 469 bp of the partial coat protein gene of BCMV and 525 bp corresponds to the CABMV coat protein. One step RT-PCR amplification of viral RNA strands were carried out. Amplification of BCMV RNA was done using 1 cycle of 44 °C for 30 min, 1 cycle of 95 °C for 1 min; 35 cycles of 94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min, 72 °C for 7 min as final extension and store forever at 4 °C. For CABMV amplification was carried out by 1 cycle of 42 °C for 30 min, 1 cycle of 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s, 72 °C for 5 min as final extension and store forever at 4 °C. The amplification products were resolved on 1.5% agarose gel stained with GR green (5 µl/100 ml) and run in the electrophoretic tank containing TAE buffer pH 8.0 at 120 V for 40 min. The gel result was viewed under a UV transilluminator (EZ imager, Bio-Rad, Inc, USA).
Data analysis
Data collected on viral disease incidence and severity were subjected to analysis of variance using SAS software with means separated by Duncan Multiple Range test at p ≤ 0.05.
Data availability
Data and materials are disclosed to manuscript. Further information on data and materials can be directed to the corresponding authors.
Abbreviations
- WAP:
-
Weeks after planting
- ACP-ELISA:
-
Antigen–coated plate enzyme linked immunosorbent assay
- AYB:
-
African yam bean
- IITA:
-
International Institute of Tropical Agriculture (IITA)
- AUDPC:
-
Area under disease progressive curve
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- CTAB:
-
Cetyltrimethyl ammonium bromide
- CPMMV:
-
Cowpea mild mottle virus
- CMV:
-
Cowpea mottle virus
- SBMV:
-
Southern bean mosaic virus
- CPMoV:
-
Cowpea mottle virus
- BCMV:
-
Bean common mosaic virus
- CABMV:
-
Cowpea Aphid-borne mosaic virus
- TSs:
-
Tropical Sphenostylis stenocarpa
- DI:
-
Disease incidence
References
Asoiro, F. U. & Ani, A. O. Determination of some physical properties of African yam beans. Pac. J. Sci. Technol. 12, 374–380 (2011).
Chinedu, S. N. & Nwinyi, C. O. Proximate analysis of Sphenostylis stenocarpa and Voadzeia subterranean consumed in South-Eastern Nigeria. J. Agric. Biotechnol. Sustain. Dev. 4, 1–6 (2012).
Ojuederie, O. B., Balogun, M. O., Fawole, I., Igwe, D. O. & Olowolafe, M. O. Assessment of the genetic diversity of African yam bean (Sphenostylis stenocarpa Hochst ex. A Rich. Harms) accessions using amplified fragment length polymorphism (AFLP) markers. Afr. J. Biotechnol. https://doi.org/10.5897/AJB2014.13734 (2014).
Saka, J., Adeniyan, O., Akande, S. & Balogun, M. An economic evaluation of intercropping African yam bean, kenaf and maize in the rain forest zone of Nigeria. Middle-East J. Sci. Res. 2, 01–08 (2007).
Kehinde, A., Obun, C., Inuwa, M. & Bobadoye, O. Growth performance, haematological and serum biochemical indices of cockerel chicks fed ginger (Zingiber officinale) additive in diets. Anim. Res. Int. 8, 1398–1404 (2011).
Hema, M., Sreenivasulu, P., Patil, B. L., Kumar, P. L. & Reddy, D. V. Tropical food legumes: Virus diseases of economic importance and their control. In Advances in Virus Research (eds Hema, M. et al.) 431–505 (Elsevier, 2014).
Amusa, N., Adigbite, A., Muhammed, S. & Baiyewu, R. Yam diseases and its management in Nigeria. Afr. J. Biotech. 2, 497–502 (2003).
Ameh, G. & Okezie, C. Pests and diseases of African yam bean, Sphenostylis stenocarpa (Hoechst. ex. A. Rich) harms. Bio-Research 3, 14–20 (2005).
Ogunsanya, O., Afolabi, C., Otusanya, M. & Adebisi, M. Responses of African yam bean (Sphenostylis stenocarpa [Hochst. Ex A. Rich]) accessions to viral diseases and serological identification of the associated viruses. Niger. J. Biotechnol. 37, 85–93 (2020).
Jones, R. A. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 141, 113–130 (2009).
Elena, S. F., Fraile, A. & García-Arenal, F. Evolution and emergence of plant viruses. Adv. Virus Res. 88, 161–191 (2014).
Akinjogunla, O. Effects of single and mixed inoculation with viruses on symptomatology, growth, yield and nutritive content of cowpea:(Vigna unquiculata). M. Sc. Thesis, University of Lagos (2005).
Aliyu, T., Balogun, O. & Kumar, L. Survey of the symptoms and viruses associated with cowpea (Vigna unguiculata (L).) in the Agroecological zones of Kwara State, Nigeria. Ethiop. J. Environ. Stud. Manag. 5, 613–619 (2012).
Amayo, R. et al. Prevalence of viruses infecting cowpea in Uganda and their molecular detection. Afr. J. Biotechnol. 11, 14132–14139 (2012).
Morales, F. J. Common beans. In Natural Resistance Mechanisms of Plants to Viruses (eds Loebenstein, G. & Carr, J. P.) 367–382 (Springer Netherlands, 2006).
Hughes, J. and Shoyinka, S. Overview of viruses of legumes other than groundnut in Africa (2003).
Kareem, K. & Taiwo, M. Interactions of viruses in cowpea: Effects on growth and yield parameters. Virol. J. 4, 1–7 (2007).
Taiwo, M. & Akinjogunla, O. Cowpea viruses: Quantitative and qualitative effects of single and mixed viral infections. Afr. J. Biotechnol. 5, 1749–1756 (2006).
Karim, F. A. Survey of cowpea viral disease symptoms and detection of associated viruses in selected cowpea growing areas in Ghana. Kwame Nkrumah University of Science and Technology (2016).
Campbell, C. L. & Madden, L. V. Introduction to Plant Disease Epidemiology (Wiley, 1990).
Kumar, P. L. Methods for the diagnosis of plant virus diseases: Laboratory manual. Int. Inst. Trop. Agric. (IITA), Ibadan, Nigeria. 15–18 (2009).
Ruiz-García, A. B., Bester, R., Olmos, A. & Maree, H. J. Bioinformatic tools and genome analysis of Citrus tristeza virus. In Citrus Tristeza Virus: Methods and Protocols (eds Catara, A. F. et al.) 163–178 (Springer New York, 2019).
Acknowledgements
The authors wish to appreciate Federal University of Agriculture Abeokuta (FUNAAB). The management and staff of the Genetic Resources Center (GRC) International Institute of Tropical Agriculture (IITA), for the fund and Germplasm Health Unit, Virology and Molecular Diagnostic Unit, for the use of the laboratory we are also deeply grateful to the assistants of staff who helped with field and laboratory work.
Funding
This research work was supported by the Genetic Resources Center (GRC), International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria, with the funds received from the CGIAR Trust Fund Donors, through the CGIAR Initiatives on Genebanks and Plant Health.
Author information
Authors and Affiliations
Contributions
J.U, I.A, E.I, M.T and O.A conceived and designed the study. J.U conducted the experiment. T.T analyzed the data P.L, I.A, E.I, M.T, T.T, P.O and O.A reviewed and edited the manuscript. P.L, I.A, and O.A reviewed and finalized the manuscript. All authors contributed to the final version of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jeffrey, I., Kehinde, I., Ayo-John, E. et al. Serological and RT-PCR evaluation of African yam bean (Sphenostylis stenocarpa (Hochst ex. A. Rich) Harms) accessions to viral resistance under field condition. Sci Rep 14, 9708 (2024). https://doi.org/10.1038/s41598-024-59977-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59977-6
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.