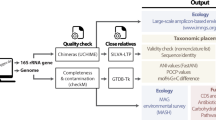
Abstract
Although the 16S rRNA gene is frequently used as a phylogenetic marker in analysis of environmental DNA, this marker often fails to distinguish closely related species, including those in the genus Vibrio. Here, we investigate whether inclusion and analysis of 23S rRNA sequence can help overcome the intrinsic weaknesses of 16S rRNA analyses for the differentiation of Vibrio species. We construct a maximum likelihood 16S rRNA gene tree to assess the use of this gene to identify clades of Vibrio species. Within the 16S rRNA tree, we identify the putative informative bases responsible for polyphyly, and demonstrate the association of these positions with tree topology. We demonstrate that concatenation of 16S and 23S rRNA genes increases the number of informative nucleotide positions, thereby overcoming ambiguities in 16S rRNA-based phylogenetic reconstructions. Finally, we experimentally demonstrate that this approach considerably improves the differentiation and identification of Vibrio species in environmental samples.
Similar content being viewed by others
Introduction
The ubiquitous presence and diversity of microorganisms make them useful indicators of environmental changes that affect the integrity and functioning of aquatic ecosystems1. These systems currently experience substantial impacts in response to ongoing climate change caused by anthropogenic emission of greenhouse gases2. Among the bacteria used as environmental indicators in monitoring aquatic environments, species in the bacterial genus Vibrio are of particular interest due to their varied and important ecological roles and impacts3,4. Besides their involvement in nutrient cycling5,6,7, a number of Vibrio species are pathogens of humans (e.g., V. cholerae, V. parahaemolyticus, V. vulnificus)8,9 and of animals10. Vibrios can cause coral bleaching (V. coralliilyticus, V. mediterranei)11, bivalve mollusc mortality12, and a variety of lesions in crustaceans and fish13,14. Further, Vibrio-associated diseases are notable in the context of climate change, which has led to an increase in Vibrio outbreaks15,16 that have caused morbidity in humans15 and substantial economic losses in the seafood industry, especially in shrimp and fish aquaculture14,17,18. The increasing incidence of infection and losses involving Vibrio, the emergence of multi-drug resistant variants13,14, and economic and public health impacts, underscore the importance of rigorous environmental monitoring of these microorganisms.
As many marine and aquatic vibrios can be cultured in vitro, and a rough assessment of Vibrio community composition and dynamics can be carried out by using culture-based techniques (e.g., incubation on selective media such as TCBS agar or CHROMagar Vibrio). However, more complete information about community structure that includes both culturable and non-culturable (dormant) vibrios is obtainable with molecular tools such as CARD-FISH19 and analysis of metagenomic DNA extracted from environmental samples. Moreover, the culturable vibrios can subsequently be purified and genotyped to reveal species identities. In particular, comparative analysis of 16S rRNA gene sequences from the ribosomal operon, along with the use of other molecular tools (e.g., DNA-DNA hybridization; DDH and reverse transcription-polymerase chain reaction, RT-PCR) and phenotypic data, have promoted the accumulation of millions of ribosomal gene sequences in reference databases such as SILVA20 and rrnDB21. These archived sequences are currently used for species identification of vibrios and other bacteria8,22. Alternatively, the high genetic diversity and number of bacterial ‘housekeeping’ genes23 have made it possible to differentiate congeneric species, and to generate phylogenetic hypotheses based on comparative analysis, also known as Multilocus Sequence Analysis (MLSA)24,25. For some time MLSA has been used to differentiate Vibrio species and conduct taxonomic assignment of Vibrio isolates26,27,28. However, except for 16S rRNA, the use of house-keeping genes as phylogenetic markers is less common in analysis of more complex environmental DNA samples (eDNA), which contain the genetic material of multiple species. The complexity and multi-species composition of eDNA makes it difficult to ensure that chimeric genomes are not formed during sequence assembly, producing artificial genotypes. Further, the use of sequence from 16S rRNA alone results in incomplete species coverage during sequencing, as well as taxonomic assignment with limited accuracy26,27. Previous studies reveal that within-genome heterogeneity of ribosomal operons draws into question species discrimination based on the sequence of a single copy of the 16S rRNA gene29,30. For instance, 16S rRNA sequences from Scytonema hyalinum strains are extremely heterogenous as previously shown31, with an intra-genomic sequence variability of 7.3–9.0%, and their use frequently leads to incorrect taxonomic assignment. Similarly, the inconsistent differentiation of the genera Butyrivibrio and Pseudobutyrivibrio32 further demonstrates limitations of 16S RNA analysis. These findings highlight some intrinsic weaknesses of 16S rRNA as a phylogenetic marker. To overcome them, Martijn et al.33 employ the 16S and 23S rRNA genes to study bacterial and archaeal diversity in environmental samples and demonstrate higher statistical support and increased number of monophyletic groups in comparison to those obtained in phylogenetic analysis of variation at single genes. Nonetheless, the efficiency of this approach for species level differentiation in the Vibrio genus has not previously been studied. Likewise, an alternative approach using the ITS regions within 16S-23S rRNA has not provided sufficient resolution to unambiguously differentiate Vibrio species in earlier work34,35.
Here, we (i) define the key sequence features that limit the potential of 16S rRNA gene to serve as a phylogenetic marker in discrimination of Vibrio species and (ii) assess the capacity of 23S rRNA to improve species resolution in phylogenetic analysis. First, we reconstruct a phylogenetic tree by using 16S rRNA sequences retrieved from 40 completely sequenced Vibrio genomes. We evaluate whether nucleotide polymorphism within 16S rRNA loci of single Vibrio species genomes might cause polyphyly and taxonomic ambiguity. Further, we identify how variation at particular nucleotide positions in 16S rRNA gene can drive the formation of polyphyletic clades during phylogeny reconstruction, and demonstrate the role of these positions in determining tree topology. We show that concatenation of 16S and 23S rRNA genes increases the number of informative nucleotide positions, thereby overcoming ambiguities in 16S rRNA-based phylogenetic reconstructions and improving the differentiation of Vibrio species. We use these results to design and test Vibrio-specific PCR primers that target the conserved terminal regions of 16S and 23S rRNA genes in order to amplify complete 16S-23S regions. Our results will help to improve the detection and identification of Vibrio species in eDNA samples, thus facilitating Vibrio monitoring in aquatic ecosystems.
Results
Through phylogenetic analysis of sequence variation among gene copies, we visualize the genetic variation within and among species that is represented in topological variation in phylogenetic trees. To minimize errors, our analysis was carried out with 40 Vibrio genomes selected based on (i) their completeness, and availability of high-quality sequencing data and annotation, along with (ii) preferentially unambiguous taxonomic assignment. The selected high-quality genomes belong to the groups that satisfy one of three levels of certainty. The first group include genomes that meet literature support and satisfy the NCBI taxonomic criteria check. The second group is limited to those that satisfy only NCBI taxonomic check. The third group contains those genomes that are not supported by any of the above criteria.
16S rRNA gene-based tree
The topological analysis of 16S rRNA gene-based tree indicates that 16S rRNA gene copies of 26 species (e.g., V. cholerae, V. vulnificus, V. casei) form monophyletic clades (MCs), indicated on the tree by triangles (Fig. 1). Unlike a “cluster” that is usually referred to a group of sequences that bare resemblance to each other regardless of their evolutionary relationship, the phylogenetic term “clade” unites the group of sequences that belong to the organisms possessing a common ancestor. Our results demonstrate that 19 MCs are highly supported by bootstrap values ≥ 95 (Fig. 1), whereas other monophyletic clades, such as V. parahaemolyticus (bootstrap equal to 54) and V. furnissii (bootstrap equal to 70), are not sufficiently supported to fully rely on this monophyletic clade formation, based on variation at the 16S rRNA locus. The sequence variation present in the 16S rRNA copies of the remaining 14 species results in polyphyly, which is manifested by clustering of one or more 16S rRNA copies of one species with those of other species (e.g., V. chagasii in light blue and V. azureus in brown; Fig. 1). The species forming polyphyletic clades mostly coincide with the second and third level categories of certainty in genome taxonomic assignment that we defined (see Materials and Methods). Three distinct types of phylogenetic tree incongruences contribute to observed polyphyly: (i) ‘outlier’, the failure of one 16S rRNA copy to cluster with the rest of the copies from the same genome (e.g., V. chagasii in light blue, V. campbellii in orange, Fig. 1); (ii) ‘breaking’, a cluster of 16S RNA gene copies of one species is placed into what would otherwise be a MC of another species, resulting in paraphyly or polyphyly (e.g., V. cholerae—V. mimicus in light pink, V. coralliilyticus—V. tubiashii in purple, Fig. 1); (iii) ‘distinct clusters’, the appearance of two separate clusters of 16S rRNA gene sequences from the same genome (e.g., V. azureus in brown, Fig. 1).
Maximum likelihood phylogenetic tree using all 16S rRNA sequences of 40 representative Vibrio genomes. Species are highlighted if they form clades in one of the phylogenetic trees (16S or 23S rRNA-based). Clades are collapsed and labelled with the corresponding species name. The number of gene copies in each clade is shown in brackets. The bootstrap values are calculated with 1000 replicates. The genome taxonomic assignment satisfies both reliability criteria (no asterisk), satisfies only the NCBI criterion (*) or does not satisfy any of the criteria (**). Two additional sequences corresponding to Salmonella bongori (16S422) and Escherichia coli (16S423) are indicated in red as an outgroup.
Disentangling polyphyletic patterns in the 16S rRNA gene-based tree
The phylogenetic ambiguity in the 16S rRNA tree appears largely associated with outlier gene copies and broken monophyly. The former is illustrated by patterns yielded by 16S rRNA copies in V. chagasii and V. campbellii, indicated by light blue and orange, respectively (Figs. 1 and 2). These gene copies present differing placements in the 16S rRNA tree, with one gene copy positioned separately from the rest of the clade, thus representing the case of an outlier gene copy (Fig. 1). The V. chagasii outlier gene copy M has three unique regions (i.e., V1, V2 and V3; Fig. 2a). Analysis of V. chagasii sequences from SILVA SSU data repository (indicated by a single asterisk in Fig. 2) reveals that they mainly contain variable regions 2 and 3, while only V. chagasii AP619 has all three variable regions (Fig. 2a).
Maximum likelihood phylogenetic trees of V. chagasii (a, blue background) and V. campbellii (b, orange background) cases illustrating the outlier gene copies with a sister clade and the variable regions associated with outlier gene copy topology. A graphical representation of 16S rRNA gene with characteristic variable regions (V1, V2 and V3) is shown in the bottom left corner of each tree. The variable regions in blue are unique to the sequences (tree tips) featured with blue text, and variable regions highlighted with magenta are shared among sequences, indicated with both blue and magenta text. The black nucleotides inside of the variable regions are not unique to the outlier gene copy. Additional sequences with the highest homologies to the two species are from the SILVA database (*). Bootstrap support is calculated with 1000 replicates. The HKY + F + R2 substitution model is used here. Two additional sequences corresponding to Salmonella bongori (16S422) and Escherichia coli (16S423) are included as an outgroup (in red).
In comparison, the outlier gene copy E of the V. campbellii genome has a variable region, V1, that is comprised of five unique nucleotides that are absent in other gene copies of the genome. However, the general composition of the V1 region is not unique. Further analysis shows that several V. campbellii sequences from the SILVA SSU data repository (Fig. 2b, indicated in blue) contain all five variable nucleotides. Moreover, one V. campbellii sequence (6-8PIN) has two variable nucleotides in this region (Fig. 2b, in magenta).
There are five instances of the second type of polyphyly, clade breaking, in the 16S rRNA tree (Fig. 1), in which a monophyletic clade is placed inside of another, otherwise monophyletic, clade. Examples include clustering of V. cholera within V. mimicus (light pink), V. jasicida within V. owensii (yellow), V. harveyi within V. azureus (brown), V. rotiferianus within V. campbellii (orange) and V. coralliilyticus inside of V. tubiashii (purple). To gain further insight, we present two cases (i.e., V. cholerae within V. mimicus, and V. jasicida within V. owensii) in further analysis. The incongruence in V. mimicus topology (Fig. 3a) is associated with several variable positions. Specifically, nucleotides in positions 219, 839, and 847 (highlighted in blue) are present in all gene copies of V. mimicus, while nucleotides in positions 632, 847, 848, and 1036 (highlighted in orange) are unique for V. cholerae. In addition, V. mimicus operon copy E exhibits cytosine (C) in position 188, a state shared with most 16S rRNA sequences of V. cholerae, while V. mimicus gene copy A has guanine (G) in position 839, which also occurs in all gene copies of V. navarrensis.
Maximum likelihood phylogenetic trees of V. cholera–V. mimicus (a) and V. jasicida–V. owensii (b), both cases forming a clade inside of a clade of another species. Informative nucleotides responsible for the species clustering patterns, i.e., nucleotides (in orange) of collapsed clades (i.e., V. cholera and V. jasicida) breaking another species clade; nucleotides (in blue) shared among gene copies of polyphyletic clades (V. mimicus and V. owensii); nucleotides (in yellow) belonging to sister species (V. navarrensis, V. campbellii and V. neocaledonicus); nucleotides (in green) associated with additional sequence sub-cluster, are specified. Sequences sharing the same nucleotide in the same position as one of the defined groups are indicated by the same color. Letter codes that represent degenerate nucleotides are in red (i.e., Y can be C or T; S can be G or C; W can be A or T; R can be A or G). The indicated nucleotides represent substitutions with one exception (i.e., deletion of one nucleotide in position 94 in gene copy “G” of V. owensii, indicated by hyphen), whereas the blank positions correspond to conserved nucleotides omitted for simplicity. The labelling by asterisks is the same as in Fig. 1. Bootstrap support is calculated with 1000 replicates. Two additional sequences corresponding to Salmonella bongori (16S422) and Escherichia coli (16S423) are included as an outgroup in red.
Several nucleotides are associated with the characteristic pattern of V. owensii 16S rRNA loci (Fig. 3b). Nucleotides in positions 75 (G), 79 (A), 94 (A/G), 98 (T), and 102 (G), highlighted in orange, are also present in most of V. jasicida sequences. Despite the presence of guanine (G) in position 94 in six V. owensii gene copies, other V. owensii gene copies (i.e., H, K, J, B and I) have adenine (A) in this location and therefore resemble sequences of V. campbellii E and V. neocaledonicus L. Additionally, V. owensii F, D, E and L have unique nucleotides in other positions (i.e., positions 96, 857, 1019 and 1028), highlighted in green. The remaining positions are either shared among V. jasicida and V. owensii gene copies G, C, and A (placed under a common node) or shared with other sister species, V. campbellii and V. neocaledonicus.
23S rRNA phylogenetic tree
Following the 16S rRNA tree-based strategy for 23S rRNA and further examination reveals that ribosomal sequences of 32 among 40 representative Vibrio species form MCs. V. diabolicus (magenta) and V. owensii (yellow) are the only species for which monophyly is not highly supported (bootstrap values equal to 76 and 91, respectively; Fig. 4). Polyphyly of an additional eight species is determined by one or several 23S rRNA gene copies clustering separately from the rest of the clade (e.g., V. splendidus, V. crassostreae, Fig. 4).
Maximum likelihood phylogenetic tree using all 23S rRNA sequences of 40 representative Vibrio genomes. Species are highlighted if they form clades in exactly one of the phylogenetic trees (16S or 23S rRNA-based). Clades are collapsed and the corresponding species names are indicated. The number of gene copies in each clade is shown in brackets. The bootstrap values are calculated with 1000 replicates. The labelling by asterisks is the same as in Fig. 1. Two additional sequences corresponding to Salmonella bongori (23S422) and Escherichia coli (23S423) are indicated in red as an outgroup.
16S and 23S rRNA gene concatenation
The above results indicate that some sequence features, including the lack of sufficient informative bases, can limit the number of MCs, especially when phylogenetic analysis is conducted with only one of either the 16S or the 23S rRNA genes. In an attempt to overcome this limitation, we alternatively generate phylogenetic trees by using concatenated 16S and 23S rRNA gene sequences (Fig. 5). Both types of 16S and 23S rRNA gene sequence concatenation (i.e., 16S-23S and 23S-16S) produce 32 MCs, of which the same 29 species are highly supported (bootstrap ≥ 95). We also show that the use of 16S-23S concatemers increases bootstrap support for clades of two species (V. mimicus and V. chagasii) and reduces support for V. vulnificus, (i.e., 91, 88 and 84, respectively; Fig. 5a) when compared to a 23S-16S tree (i.e., 71, 42 and 93, respectively; Fig. 5b). Moreover, although V. neocaledonicus and V. alginolyticus (Fig. 5b) form a monophyletic clade in the 23S-16S tree, this clade emerges as polyphyletic in the 16S-23S tree (Fig. 5a) due to insertion of V. diabolicus and V. natriegens sequences. In contrast to the MCs formed by V. campbellii (orange) and V. owensii (yellow) on the 23S rRNA tree (Fig. 4), these species do not form MCs in the 16S-23S and 23S-16S gene concatenation trees (Fig. 5a,b). However, the use of 16S-23S concatemers instead of 23S rRNA sequences better differentiates three species (V. diabolicus, V. natriegens and V. vulnificus; Figs. 4 and 5a) that were not resolved by 23S rRNA alone.
Maximum likelihood phylogenetic trees using 16S and 23S rRNA concatenated sequences in order 5′-16S-23S-3′ rRNA (panel a) or 5′-23S-16S-3′ rRNA (panel b) of 40 representative Vibrio genomes. To simplify the view, clades are collapsed and labelled with the corresponding species name. The number of gene copies in each clade is shown in brackets. Bootstrap support is calculated with 1000 replicates. V. neocaledonicus and V. alginolyticus are shown in blue. The labelling by asterisks is the same as in Fig. 1. Additional sequences corresponding to Salmonella bongori (16S-23S422 and 23S-16S422, respectively) and Escherichia coli (16S-23S423 and 23S-16S423, respectively) are indicated in red as an outgroup.
Conserved and variable regions in Vibrio 16S and 23S rRNA genes
Since concatenation improves bootstrap support, we analyze further 16S and 23S rRNA loci to identify variable and conserved regions, and subsequently design Vibrio-specific primers for amplifying the entire 16S-23S genomic region. First, we find that the 16S rRNA locus is highly conserved across the Vibrio genus (Fig. 6). Approximately 74.6% of positions (1210) are fully conserved, whereas 162 positions are variable (Supplementary Table S1). Nine variable regions of 16S rRNA (indicated by brackets, Fig. 6) universally present in bacteria30,36 and variable regions identified in the alignment of Vibrio 16S rRNA sequences (indicated in red, Fig. 6) largely coincide. Regions 1, 3, 4, and 6 are the most variable in Vibrio, while regions 5, and 7 do not contain any variable positions except a 12-nucleotide-long insertion present in V. metschnikovii gene copy E, and guanine (G) insertion in V. rumoiensis gene copy B in variable regions 5 and 7, respectively (Fig. 6). Regions 2, 8, and 9 have some variable positions as well.
Variable and conserved regions of 16S rRNA and the location of 16S rRNA locus-specific forward primer. Sequence alignment of 16S rRNA gene sequences that are present in representative Vibrio genomes reveals the positions of variable (in red), fully conserved (dark gray), and conserved (in light gray) regions. The gaps are shown as white areas. Brackets indicate variable regions universally present in bacteria30,36. The position of 16S rRNA gene-specific forward (F) primer is shown at the bottom.
Second, our approximate delimitation of conserved and variable regions of Vibrio 23S rRNA sequences retrieved from SILVA LSU r138.1 RefNR data repository (Fig. 7a) reveals that 57.43% (1743) of the positions are fully conserved (Supplementary Table S1). By allowing a single mismatch in one of the aligned sequences per position, the conserved region can approximately cover 73.34% of the total alignment. In further interpretation of 303 variable positions (represented by red bars, Fig. 7a) we define variable regions as those with high concentration of variable nucleotide positions, i.e., those at which the consensus nucleotide occurs at frequencies lower than 0.747 (see Materials and Methods section and Fig. 7a). We find ten variable regions in aligned Vibrio 23S rRNA gene sequences. Moreover, variable region 3 is split into two subregions (i.e., 3a and 3b; Fig. 7a). In contrast to the alignment of 23S rRNA SILVA sequences, the 23S rRNA sequence alignment of our data repository (Fig. 7b) has a higher percentage of fully conserved regions (77.47%), which leads to a higher cutoff frequency value of 0.893 (Supplementary Table S1). The variable regions coincide in both alignments (Fig. 7), except for the presence of additional variable positions at the 3’ end (Fig. 7b). We include this additional region as V10a in Fig. 7b to differentiate shared variable positions in sequences retrieved from SILVA from those held in our repository.
Deduced variable and conserved regions of 23S rRNA based on frequency values with which the consensus nucleotide at each aligned position occurs in Vibrio sequences retrieved from SILVA database (a) and a local data repository of representative Vibrio genomes (b). Indicated are fully conserved positions (dark gray), variable (red), conserved (light gray), and gaps (white). The position of the white dots representing individual consensus nucleotides is adjusted according to the frequency occurring at each position. A blue dashed line represents the cutoff frequency separating ten percent of the most variable nucleotides.
Selection of primers for amplification of Vibrio 16S-23S region
The assessment of several universal PCR primers allows selection of candidates for forward and reverse primers for the entire 16S-23S region (including both ribosomal RNA genes and intergenic spacer) in Vibrio genomes (Fig. 6, Supplementary Tables S2 and S3). Preliminary literature review reveals a main candidate location suitable for designing universal 16S rRNA gene-specific bacterial primers. This candidate location corresponds to primer variants similar to 27F, a universal forward primer widely used to amplify 16S rRNA bacterial gene sequences37. Furthermore, comparison of these variants provides a consensus primer sequence (c27F) representing all available primer variants (Supplementary Table S2). Although the universal bacterial forward primer (i.e., S-DBact-0008-cS-20, Supplementary Table S2) containing the same number of degenerate bases as c27F allows a better coverage among bacteria than a less degenerated primer, the Vibrio-specific consensus sequence eliminates the necessity of using such a highly degenerated forward primer for Vibrio species (Supplementary Table S2). We found that all custom Vibrio 16S rRNA gene copies can be amplified by the original 27F universal primer (5’-AGAGTTTGATCMTGGCTCAG-3’) introduced in 1991 (Supplementary Table S2).
After defining conserved regions in 23S rRNA genes (Fig. 7b), we consider two conserved regions as targets for a Vibrio-specific 23S rRNA reverse primer. Among them, the sense strand target of primer 23S_rev_V (positions 2864 to 2285) is closer to the 3’end of 23S rRNA sequence than the region in the sense strand (positions 2227 to 2243) complementary to primer 2242R. The first region provides a longer conserved sequence without indels and should provide nearly full length 23S rRNA amplicons, making 23S_rev_V the best candidate as a universal Vibrio-specific primer (Supplementary Table S3).
An in silico specificity test employing the locus-specific 27F and 23S_rev_V primer pair for amplification of the 16S-23S region of complete Vibrio and non-Vibrio genomes does not yield products with non-Vibrionaceae genomes as templates (Fig. 8). In contrast, in silico PCR produces amplicons of anticipated length for all Vibrio genomes as well as for five non-Vibrio Vibrionaceae genomes (Fig. 8, Supplementary Table S4). Furthermore, the same primers amplify in silico all 16S-ITS-23S copies from 40 Vibrio genomes from our database (Fig. 8). All the amplicons obtained have the expected size, approximately 4700 bp.
The results of in silico PCR amplifications obtained by the combination of 16S rRNA 27F42 and 23S rRNA 23S_rev_V primers. Whole genome sequences of Vibrio, non-Vibrio Vibrionaceae and non-Vibrionaceae species are used as templates. Genomes for which gene copies can be amplified (shaded) and can not be amplified (blue). The lack of amplification in the case of non-Vibrionaceae genomes suggests that the sequence of Vibrio-specific primers was not conserved in these genomes. Thus, the primers demonstrate preferential specificity for Vibrionaceae family.
Experimental validation of Vibrio-specific primers
To experimentally test the ability of the Vibrio-specific primers to generate 16S-ITS-23S amplicons, we extracted metagenomic DNA from an environmental water sample and used it as a template. Consistent with the in silico results (see previous section), PCR amplification of environmental DNA using 27F and 23S_rev_V primers yields amplicons of the expected size (Supplementary Fig. S1). Their sequencing with Oxford Nanopore technology and standard protocols produced 105,230 reads (see “Materials and methods”; Supplementary Table S5). Analysis of high quality reads (83,207 in total) by WIMP (Oxford Nanopore) revealed that almost all reads belong to Proteobacteria (see Supplementary Fig. S2). Moreover, nearly half of these reads (48.9%) are of Gammaproteobacteria origin. We show that 2.17% reads correspond to the species that belong to the Vibrionaceae family (Supplementary Table S5). These reads (1,806 in total) represent a large variety of Vibrio species (see Supplementary Table S6). Interestingly, the trimming of 16S-ITS-23S reads at their 3’ end to obtain 1600 nt fragments (representing 16S rRNA gene sequences) and subsequent taxonomic annotation by WIMP (Oxford Nanopore) demonstrates that nearly one third of the truncated reads (i.e., 33.9%) become assigned to non-Vibrio species (Supplementary Fig. S2, panel c). Moreover, the trimming-dependent “loss” of some Vibrio reads also decreases the number of species compared to that initially discovered based on 16S-ITS-23S sequences (Supplementary Table S6).
Discussion
Improved differentiation of Vibrio species has been possible using the 23S rRNA gene as a phylogenetic marker instead of the 16S rRNA gene. Analysis of a maximum likelihood (ML) 16S rRNA gene tree identifies the informative bases associated with multiple polyphyletic patterns (Figs. 2 and 3), which are largely resolved in a 23S rRNA gene tree. The 23S rRNA gene tree presents 11 additional highly supported monophyletic clades compared to the 16S rRNA gene tree (Figs. 1 and 4). The capacity of a 23S rRNA gene tree to reveal a higher number of monophyletic clades than a 16S rRNA gene phylogeny is consistent with the results obtained for non-Vibrio taxa38. The higher number of informative bases within the 23S rRNA gene when compared to 16S rRNA (i.e., 295 vs 162 variable positions; Supplementary Table S1) likely accounts for the observed increase in differentiation of Vibrio and non-Vibrio species.
We combine 16S rRNA and 23S rRNA gene sequences to increase the number of informative bases, in order to distinguish additional Vibrio species. The 16S-23S concatemer-based trees form a number of monophyletic clades similar to that provided by the 23S rRNA tree (Figs. 4 and 5, respectively), thus resembling the results for non-Vibrio species that are obtained with trees of single copies (per genome) of 23S rRNA and 16S-23S concatemers38. Nonetheless, the trees based on the concatemer sequences (Fig. 5) enable taxonomic assignment of three additional Vibrio species (V. diabolicus, V. natriegens and V. vulnificus). These species are not resolved in the phylogenetic trees based on Vibrio 23S rRNA or 16S rRNA sequences individually.
A new combination of 16S forward and 23S reverse primers for in silico amplification of Vibrio 16S-23S region suggests the feasibility of targeting Vibrio-specific sequences in environmental DNA. Amplification of this region in bacteria usually involves universal 16S rRNA 27F and 23S rRNA 2490R39,40 or 2241R41 primers. To increase the specificity of amplification, assure the broadest coverage of Vibrio species, and produce amplicons with an increased number of informative bases, we propose the combination of forward (16S rRNA 27F42) and new reverse (23S rRNA 23S_rev_V) primers. These primers can amplify in silico the corresponding fragments of all Vibrio ribosome operons in our custom database and provide products of 4.7 kbp or more. This size matches that of amplicons (i.e., 4.3–5.4 kbp) that encompass the entire 16S-ITS-23S regions in a large variety of bacterial species41.
Intragenomic variability in the number of rRNA operons constrains the use of ribosomal genes for analysis of environmental samples (https://rrndb.umms.med.umich.edu/search/)21. We observe that the number of ribosomal operons ranges from 5 to 16 for Vibrio genomes (Supplementary Fig. S3), and is on average higher than that reported in other studies11,35. Ribosomal operon multiplicity allows bacteria to increase ribosomal content quickly, providing rapid adaptation to changing environmental conditions, such as increase in nutrient availability or favorable temperature shifts43. Intragenomic variability in number of operons among strains that belong to the same species could lead to over- or underestimation of species richness, as the real number of species in samples can be lower/higher than that estimated based on the number of detected unique gene copies30. This circumstance restricts the application of ribosomal genes in quantitative analysis of eDNA44.
The general plasticity of bacterial genomes extends to the nucleotide composition of ribosomal operons, and therefore can influence outcomes of phylogenetic analysis. The presence of 16S rRNA and 23S rRNA in the same molecular machine (i.e. the ribosome) suggests their interdependent evolution to preserve ribosomal function. Compared to sequence conservation in regions important for ribosomal function, variable regions show higher diversity and are the location of informative bases in phylogenetic analysis. The phylogenies of our 16S rRNA and 23S rRNA gene copies from the same operon reveal different evolutionary relationships for some Vibrio species (Fig. 1 vs Fig. 4). These observations suggest that the routine sequencing of 16S rRNA amplicons and subsequent homology search can produce false matches for “outlier gene copies”, and consequently mislead species assignment. In this context, species assignment will benefit from fully annotated genomes based on all ribosomal operons30,44,45. The divergence of ribosomal operons can be explained by horizontal gene transfer or effects of mutation46,47. Even though gene transfer and mutations may increase intragenomic operon divergence in Vibrio, many studies report selection that favors homogeneous ribosomal structure and maintenance of function47,48. Intragenomic operon divergence may be transitory, and may provide an opportunity to study processes of operon homogenization.
Internal transcribed spacer (ITS; Supplementary Fig. S4) represents an additional source of informative bases contributing to the variability of the Vibrio ribosomal operon. Previous studies support the potential of ITS as a phylogenetic marker for differentiation of bacterial taxa from distinct families39,41. However, the effectiveness of ITS alone as a marker in a narrower range of taxa, such as species in the genus Vibrio, likely decreases. Consistent with this idea, the results of a previous study indicate that the use of ITS alone is insufficient for differentiating all Vibrio species35. Furthermore, lower intergenomic than intragenomic ITS sequence variability33 can further complicate species delineation based on ITS as a single marker. Despite some apparent limitations of ITS use, this region in combination with 16S and 23S rRNA genes could increase the total number of informative bases available for phylogeny construction and, therefore, might further improve the taxonomic assignment of Vibrio species.
We show through in silico analysis that, despite the key role of 16S rRNA gene in establishing the taxonomy of bacterial species, this gene possesses a number of deficiencies that complicate its use for differentiating Vibrio species in multispecies assemblages, such as those in environmental samples. Moreover, we demonstrate that some limitations can be overcome by the joint use of 16S and 23S rRNA genes, and we propose a candidate universal primer pair for Vibrio-specific amplification of the rRNA genes and the ITS. Although the joint use of ribosomal genes per se does not allow delineating all Vibrio species, the additional incorporation of ITS sequences present in the amplified 16S-ITS-23S fragments may increase the number of informative bases, potentially providing further improvements in the differentiation of Vibrio species in environmental samples.
The experimental testing of the proposed primers reveals that they work well with environmental DNA and are capable of amplifying a wide range of Vibrio sequences. Moreover, the use of these primers makes it possible to greatly increase the discoverability of Vibrio species compared to the “classical” 16S rRNA-based approaches widely used to monitor microbial diversity. For instance, one of the previous studies49 using 16S rRNA gene along as a phylogenetic marker apparently failed to identify any member of the Vibrionaceae family in environmental samples obtained from the same area (Plentzia Bay). Finally, 35.4% of Vibrio sequences that we discovered in the environmental sample originate from genomes that are not present among the genomes of 40 Vibrio species we initially selected for analysis. This strongly suggests that our primers enable broad coverage of Vibrio species.
Materials and methods
Creating a custom repository of 16S and 23S rRNA gene sequences
Vibrio is the most diverse genus in Vibrionaceae, currently including 151 described species and 5 subspecies (LPSN database, https://www.bacterio.net/, accessed June 2022)50. To carry out in silico analysis, we created a data repository by retrieving all copies of ribosomal operon genes (i.e., 16S rRNA and 23S rRNA) from 40 representative, fully-sequenced Vibrio genomes, one genome per species (Supplementary Table S7). Genome taxonomic assignment was further verified when Vibrio spp. didn’t form highly supported and unambiguously differentiated monophyletic clades. We classified levels of certainty of genome taxonomic assignment in the following way: first, literature support existed and the NCBI taxonomic check criteria were satisfied; second, only the NCBI taxonomic check criteria were satisfied; and third, when none of these criteria were satisfied (Fig. 1, Supplementary Table S8). When multiple genomes were available, we preferentially selected published and annotated genomes of validated Vibrio species in the LPSN database that were assembled using both long- and short-read sequences (e.g., those obtained by both PacBio and Illumina sequencing). To choose representative genomes of V. diabolicus, V. natriegens, and V. scophthalmi from IMG/M database (https://img.jgi.doe.gov/cgi-bin/m/main.cgi), we constructed a similarity matrix of gene copies from the same genome based on NCBI BLASTn results (https://blast.ncbi.nlm.nih.gov/) and analyzed the number of gaps and mismatches to find the genomes with the highest internal variability in 16S and 23S rRNA gene copies. Next, the ribosomal sequences that were downloaded from NCBI GenBank and IMG/M databases (Supplementary Fig. S5) were manually curated by adding missing conserved terminal nucleotides to obtain full-length copies. We assigned to each retrieved sequence a unique ID in which the last three digits referred to the operon carrying the corresponding 16S and 23S rRNA gene copies and a letter to distinguish each operon within the corresponding genome. We employed our custom code (Parts 1–5, see supplementary file “Custom code”) based on the automated webpage scraping functionality in the RSelenium (Version 1.7.7)51 and rEntrez packages (Version 1.2.2)52 to formulate a search query in R (Version 1.1.442) to obtain species and strain names, sequence accession numbers, and the corresponding sequences in FASTA format.
We obtained additional 16S rRNA sequences from the SILVA SSU r.138.1 database20. We used these sequences to ascertain whether outlier gene copies were fortuitous and potentially caused by sequencing errors, or occur more broadly in a larger sample of sequenced genes. We conducted a BLASTn homology search with the variable regions of outlier gene copies V. chagasii M and V. campbellii E. We subsequently used the five SILVA sequences with complete 16S rRNA sequence and the highest BLAST homology in polyphyly analysis of the 16S rRNA-based phylogenetic tree (Fig. 2). Additionally, 2072 non-redundant Vibrio 23S rRNA sequences were also retrieved from SILVA LSU Ref NR r.138.1 database20, corresponding to 45 species and 19 additional strains without species designation. These were then used to locate 23S rRNA conserved regions for PCR primer design (Fig. 7a, Supplementary Fig. S5). We further supplemented our repository with 26 genomes that belong to non-Vibrio species in Vibrionaceae and nine other non-Vibrionaceae bacteria. The non-Vibrio Vibrionaceae genera included Aliivibrio, Photobacterium, Salinivibrio, Enterovibrio and Grimontia, whereas non-Vibrionaceae families included Woeseiaceae, Comamonadaceae, Rhodobacteraceae, Desulfobacteraceae and Enterobacteriaceae (Escherichia coli).
Alignment, curation, trimming and concatenation of rRNA gene sequences
We used MAFFT (Version 7.490) with a global strategy (G-INS-I)53 for sequence alignment. The MAFFT algorithm provided better-aligned sequences than those obtained by using other popular algorithms such as MUSCLE and ClustalW, based on inspection. After alignment, several incomplete and apparently misannotated 16S and 23S rRNA sequences were identified and subsequently curated using the full genome sequences previously retrieved from NCBI GenBank in order to assure a standard, full-length representation of gene sequences (Supplementary Fig. S5). Moreover, some aligned sequences were manually trimmed in MEGA-X at their 5’- and 3’ extremities to ensure uniform length of sequences flanking the conserved rRNA regions across all aligned sequences.
Phylogenetic reconstruction
We used MAFFT to align and MEGA-X54 to curate rRNA gene sequences and the fusion variants (i.e., 5’-16S-23S-3’ and 5’-23S-16S-3’) obtained by concatenation of 16S and 23S rRNA from the same operon. These were then used to construct phylogenetic trees. Tree construction was performed in IQTREE (Version 2.1.3)55 at operon resolution using ML. The IQTREE algorithm automatically chose the best nucleotide substitution model for each case by selecting the model with the lowest Bayesian Information Criterion value (i.e., TIM3 + F + R5 for 16S rRNA, GTR + F + R7 for 23S rRNA, GTR + F + R7 for 16S-23S, and GTR + F + R6 for 23S-16S rRNA concatemers, respectively). IQTREE employs bootstrapping to describe node support in the reconstructed trees. We considered a node highly supported when the bootstrap value was ≥ 9556. We visualized the NEWICK format output file (.treefile) using the online tree editing program iTOL (Version 6, https://itol.embl.de/upload.cgi)57. We edited trees for better presentation using the vector graphics program Inkscape (Version 1.0, https://inkscape.org/release/inkscape-1.0/).
To interpret ambiguous sequence topologies in the 16S rRNA tree of the V. chagasii gene copy M and V. campbellii gene copy E (outlier gene copies), we visually identified the regions that are associated with differences between these outlier gene copies and other gene copies from the same genome. To assure that the unique sequence composition defining these regions was not artifactual (e.g., sequencing error), we checked for nucleotide conservation among other sequences of the same species by performing BLASTn search (https://blast.ncbi.nlm.nih.gov/) of these unique regions in additional V. chagasii and V. campbellii sequences from the SILVA SSU r.138.1 database20. Further, we employed Gblocks software58 to compare aligned 16S rRNA sequences of Vibrio spp. that form monophyletic clades that disrupt otherwise monophyletic Vibrio spp. clades (e.g., V. cholera–V. mimicus, V. jasicida–V. owensii, V. harveyi–V. azureus, V. rotiferianus–V.campbellii, and V. coralliilyticus–V. tubiashii). We refer to these cases as “broken” monophyly. Among the variable positions highlighted by Gblocks, we identified by inspection the informative nucleotides (i.e., nucleotide variants in some positions of 16S rRNA that were shared with closely related Vibrio species) that were associated with a particular topology in the phylogenetic tree. The IQ-TREE output file of the full 16S rRNA phylogenetic tree was pruned to point to a node with a distinct topology with regard to its polyphyletic sister clade and an outgroup.
Variable region identification
To identify variable regions, we first used Biostrings R package (Version 2.64.0)59 employing our custom code (Part 6, see supplementary file “Custom code”) to score the sequence conservation for each nucleotide present in the curated and trimmed alignment of rRNA gene sequences. The consensus sequence was determined by the highest frequency base at each nucleotide position. Later, positions were sorted by these values. We considered variable positions to be the ten percent of positions with the lowest frequency values, x (i.e., x < 0.768 for 16S rRNA, x < 0.893 for representative Vibrio 23S rRNA and x < 0.747 for SILVA 23S rRNA sequences), while the remaining positions were referred to as partly conserved (0.768 ≤ x < 1, 0.893 ≤ x < 1, and 0.747 ≤ x < 1 for 16S rRNA and 23S rRNA representative Vibrio and SILVA sequences, respectively) or highly conserved (x = 1). Finally, we visually identified variable regions as containing a relatively high frequency of variable positions. The location of the variable regions was further corrected and refined based on comparison with the 16S variable regions reported in other studies, as well as through analysis of a larger set of Vibrio sequences available in SILVA LSU Ref NR database for 23S rRNA.
Design of primers for amplification of the full-length 16S-23S regions of Vibrio genomes and their in silico testing
We assessed the degree to which 10 forward 16S rRNA gene-specific primers (Supplementary Table S2), previously used to amplify 16S rRNA genes, successfully hybridize with Vibrio 16S rRNA gene sequences. Among the primers that hybridize Vibrio 16S rRNA sequences, 27F primer, with a single degenerate base, provided sufficient coverage of Vibrio sequences.
To design 23S rRNA gene-specific primers, the longest conserved regions at the 3’ termini of aligned 23S rRNA sequences of representative Vibrio species were chosen as potential targets for new primers (Supplementary Table S3). To ensure base pairing in the variable positions, we allowed for primers with degenerate bases (e.g., Y represents C or T; R represents A or G; H represents A, C or T). We also included two universal 23S rRNA bacterial primers that target an internal region of this gene to assess the ability of these primers to amplify Vibrio sequences in our data repository. The location of primer binding was assessed using BLAST and the Vibrio sequences in the SILVA LSU r138.1 Ref NR database.
We tested the suitability of two primers, the 16S rRNA gene-specific 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 23S rRNA gene-specific 23S_rev_V (5’-TARRHCTCAYGGGYRATTAGTR- 3’), to serve as universal primers for amplification of nearly full-length 16S-23S region, using the in silico PCR Experiment Simulation System (Ipcress; 2.2.0 exonerate, glib version 2.47.0). The amplification targeted sequences in our custom repository of Vibrio, non-Vibrio and non-Vibrio Vibrionaceae genomes, and we specified conditions that allowed up to three nucleotide mismatches and set up the upper limit for the length of amplicons (i.e., 6000 nt).
Water sample processing and extraction of metagenomic DNA
The environmental water sample (ES_Ple_Mar) was collected in the Estuary of Plentzia in March of 2023. One and a half liter of water collected from the surface and prefiltered through a 200 μm mesh was sequentially filtered through a 3 μm polycarbonate filters (142mm diameter) followed by a 0.22 μm Sterivex™ filter unit (Millipore) using a MasterFlex Easy-Load peristaltic pump. The sterivex filter with the attached biological material was further used to extract metagenomic DNA following the DNeasy PowerWater Sterivex Kit (Qiagen) protocol. The concentration of the extracted DNA (25.8 ng/μL) was determined using a Qubit 4 Fluorometer (Thermo Fisher Scientific).
PCR amplification and gel purification of 16S-ITS-23S amplicons
The standard mixtures (50 μL) used to carry out PCR contained 19 μL of molecular biology grade water, 2.5 μL of forward primer (20 pmol/μL), 2.5 μL of reverse primer (20 pmol/μL), 1 μL of template DNA and 25 μL of 2 × Platinum SuperFi II Green PCR Master Mix (Thermo Fisher Scientific). PCR was performed by using a Veriti Thermal Cycler (Applied Biosystems, USA). The amplification process included an initial denaturation step (30 s, 98 °C) and 35 cycles of amplification (denaturation for 10 s at 98 °C, annealing for 10 s at 60 °C and extension for 2.5 min at 72 °C) followed by a final incubation for 5 min at 72 °C. The products of PCR amplification contained in two PCR tubes (100 μL in total) were deproteinized by extraction with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1). Then, an aliquote of the deproteinazed sample (6 μL) along with Gene Ruler 1kb Plus DNA Ladder (Fisher Scientific, USA) were further analysed by electrophoresis in an 1% agarose gel, followed by staining with GelRed (Millipore) and destaining in distilled water. The image of the destained gel was captured by using a ChemiDoc imaging system (Bio-Rad). To increase the yield of the target PCR product, it was reamplified using the deproteinized amplicon as a template. Aliquotes of the amplified products (50 μL each) were individually mixed with 10 μL of ROTI®Load DNAstain 2 SYBR® Green (Carl Roth) and were subsequently fractionated on a 1% agarose gel in triplicate. The DNA fragments of appr. 4500–5000 bp were visualized using a Large Blue LED Transilluminator (IO Rodeo) and extracted from the gel using the GeneJET Gel Extraction Kit (Thermo Scientific). The concentration of the extracted DNA (69.52 ng/μL) was determined using a Qubit 4 Fluorometer (Thermo Fisher Scientific).
Oxford Nanopore sequencing and post-sequencing data processing
Purified 16S-ITS-23S amplicons were further used for preparation of library conducted following the Native Barcoding Kit 24 V14 protocol instructions (SQK-NBD114-24. Oxford Nanopore Technologies, ONT, Oxford, UK). The resulting library was loaded to the MinION Mk1B flow cell FLO-MIN114 for sequencing. The obtained reads were basecalled by Dorado basecaller, installed in MinKNOW, employing the High-accuracy basecalling model, 400bps-5 kHz chemistry and the default minimum quality score (Qscore = 9). All 16S-ITS-23S reads above Qscore nine were automatically grouped by MinKNOW in fastq files. The taxonomic classification and quantitative analysis of these reads was performed by EPI2ME Desktop Agent version 3.7.3 using “What’s in my pot” workflow (WIMP 2023.06.13-1865548, ONT). This workflow analyzed reads within 4649–5538 bp length range as defined in filtering condition (Supplementary Table S5). Based on WIMP taxonomic analysis, we calculated relative abundances (%) of the identified taxa by comparing the number of reads assigned to a particular taxon and the total number of reads obtained for the sample (Supplementary Table S5, Supplementary Fig. S2). Reads identified as Homo sapiens were filtered out from fastq files. The sequences of the final reads were deposited into the public NCBI SRA database and have the following accession number: PRJNA1081186.
Data availability
The Oxford Nanopore sequencing data included and discussed in the manuscript are deposited into the NCBI SRA database as a BioProject with accession number PRJNA1081186.
Code availability
The code used for sequence scraping from online databases and nucleotide frequency calculation of aligned sequences is available as Custom code.txt file in Supplementary Material.
References
Ma, F. et al. Development of microbial indicators in ecological systems. IJERPH 19, 13888 (2022).
IPCC. IPCC, 2014: Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change [Core Writing team, R. K. Pachauri and L.A. Meyer (eds.)]. IPCC, Geneva, Switzerland, 151 pp. (2014).
Costa, R. A., Silva, G. C., Peixoto, J. R. O., Vieira, G. H. F. & Vieira, R. H. S. F. Quantification and distribution of vibrio species in water from an estuary in Ceará-Brazil impacted by shrimp farming. Braz. J. Oceanogr. 58, 183–188 (2010).
Jesser, K. J. & Noble, R. T. Vibrio ecology in the neuse river estuary, north carolina, characterized by next-generation amplicon sequencing of the gene encoding heat shock protein 60 (hsp60). Appl. Environ. Microbiol. 84, e00333-e418 (2018).
Keyhani, N. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta (BBA) Gen. Subjects 1473, 108–122 (1999).
Goecke, F., Labes, A., Wiese, J. & Imhoff, J. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–299 (2010).
Moi, I. M. et al. Polyunsaturated fatty acids in marine bacteria and strategies to enhance their production. Appl. Microbiol. Biotechnol. 102, 5811–5826 (2018).
Farmer, J. J., Michael Janda, J., Brenner, F. W., Cameron, D. N. & Birkhead, K. M. Vibrio. In: Whitman, W. B. et al. eds., Bergey’s Manual of Systematics of Archaea and Bacteria 1–79 (Wiley, 2015). https://doi.org/10.1002/9781118960608.gbm01078.
Sampaio, A., Silva, V., Poeta, P. & Aonofriesei, F. Vibrio spp.: Life strategies, ecology, and risks in a changing environment. Diversity 14, 97 (2022).
Grimes, D. J. The vibrios: Scavengers, symbionts, and pathogens from the sea. Microb. Ecol. 80, 501–506 (2020).
Rubio-Portillo, E., Yarza, P., Peñalver, C., Ramos-Esplá, A. A. & Antón, J. New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J. 8, 1794–1807 (2014).
Travers, M.-A., Boettcher Miller, K., Roque, A. & Friedman, C. S. Bacterial diseases in marine bivalves. J. Invert. Pathol. 131, 11–31 (2015).
Montánchez, I. & Kaberdin, V. R. Vibrio harveyi: A brief survey of general characteristics and recent epidemiological traits associated with climate change. Marine Environ. Res. 154, 104850 (2020).
Sanches-Fernandes, G. M. M., Sá-Correia, I. & Costa, R. Vibriosis outbreaks in aquaculture: Addressing environmental and public health concerns and preventive therapies using gilthead seabream farming as a model system. Front. Microbiol. 13, 904815 (2022).
Baker-Austin, C., Stockley, L., Rangdale, R. & Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective: V. vulnificus and V. parahaemolyticus in Europe. Environ. Microbiol. Rep. 2, 7–18 (2010).
Vezzulli, L., Colwell, R. R. & Pruzzo, C. Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65, 817–825 (2013).
Novriadi, R. Vibriosis in aquaculture. omni.akua 12, 1–12 (2016).
Mohd Yazid, S. H., Mohd Daud, H., Azmai, M. N. A., Mohamad, N. & Mohd Nor, N. Estimating the economic loss due to vibriosis in net-cage cultured Asian Seabass (Lates calcarifer): Evidence from the east coast of peninsular Malaysia. Front. Vet. Sci. 8, 644009 (2021).
Ogayar, E. et al. Efficiency and specificity of CARD-FISH probes in detection of marine vibrios. Environ. Microbiol. Rep. 13, 928–933 (2021).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K. & Schmidt, T. M. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015).
Yarza, P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014).
Gutacker, M. et al. Population genetics of Vibrio vulnificus : Identification of two divisions and a distinct eel-pathogenic clone. Appl. Environ. Microbiol. 69, 3203–3212 (2003).
Maiden, M. C. J. et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95, 3140–3145 (1998).
Glaeser, S. P. & Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 38, 237–245 (2015).
Thompson, F. L. et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71, 5107–5115 (2005).
Cano-Gomez, A., Høj, L., Owens, L. & Andreakis, N. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst. Appl. Microbiol. 34, 561–565 (2011).
Sawabe, T. et al. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 4, (2013).
Kitahara, K. & Miyazaki, K. Revisiting bacterial phylogeny: Natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mobile Genet. Elem. 3, e24210 (2013).
Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029 (2019).
Johansen, J. R. et al. Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PLoS ONE 12, e0186393 (2017).
Li, D. et al. Atypical bacterial rRNA operon structure is prevalent within the Lachnospiraceae, and use of the 16S–23S rRNA internal transcribed spacer region for the rapid identification of ruminal Butyrivibrio and Pseudobutyrivibrio strains. Ann. Microbiol. 64, 1623–1631 (2014).
Martijn, J. et al. Confident phylogenetic identification of uncultured prokaryotes through long read amplicon sequencing of the 16S-ITS-23S rRNA operon. Environ. Microbiol. 21, 2485–2498 (2019).
Hoffmann, M. et al. PCR-based method for targeting 16S–23S rRNA intergenic spacer regions among Vibrio species. BMC Microbiol. 10, 90 (2010).
Yu, J. et al. Species‐specific Identification of Vibrio sp. based on 16S‐23S rRNA gene internal transcribed spacer. J. Appl. Microbiol. 129, 738–752 (2020).
Coenye, T. & Vandamme, P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 228, 45–49 (2003).
Karst, S. M. et al. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 36, 190–195 (2018).
de Oliveira Martins, L., Page, A. J., Mather, A. E. & Charles, I. G. Taxonomic resolution of the ribosomal RNA operon in bacteria: implications for its use with long-read sequencing. NAR Genom. Bioinf. 2, lqz016 (2020).
Sabat, A. J. et al. Targeted next-generation sequencing of the 16S–23S rRNA region for culture-independent bacterial identification—increased discrimination of closely related species. Sci. Rep. 7, 3434 (2017).
Peker, N. et al. A comparison of three different bioinformatics analyses of the 16S–23S rRNA encoding region for bacterial identification. Front. Microbiol. 10, 620 (2019).
Benítez-Páez, A. & Sanz, Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinIONTM portable nanopore sequencer. GigaScience 6, (2017).
Lane, D.J. 16S/23S RRNA Sequencing in Nucleic Acid Techniques in Bacterial Systematics (Eds. Stackebrandt, E. & Goodfellow, M.) 115–175 (John Wiley and Sons, 1991).
Roller, B. R. K., Stoddard, S. F. & Schmidt, T. M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1, 16160 (2016).
Větrovský, T. & Baldrian, P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 8, e57923 (2013).
Pei, A. et al. Diversity of 23S rRNA Genes within Individual Prokaryotic Genomes. PLoS ONE 4, e5437 (2009).
González-Escalona, N., Romero, J. & Espejo, R. T. Polymorphism and gene conversion of the 16S rRNA genes in the multiple rRNA operons of Vibrio parahaemolyticus. FEMS Microbiol. Lett. 246, 213–219 (2005).
Espejo, R. T. & Plaza, N. Multiple ribosomal RNA operons in bacteria; their concerted evolution and potential consequences on the rate of evolution of their 16S rRNA. Front. Microbiol. 9, 1232 (2018).
Hassler, H. B. et al. Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies. Microbiome 10, 104 (2022).
Lanzén, A., Mendibil, I., Borja, Á. & Alonso-Sáez, L. A microbial mandala for environmental monitoring: Predicting multiple impacts on estuarine prokaryote communities of the Bay of Biscay. Mol. Ecol. 30, 2969–2987 (2021).
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C. & Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612 (2020).
Harrison, J. RSelenium: R Bindings for ‘Selenium WebDriver’. R package version 1.7.7. (2020).
Winter, D. J. rentrez: An R package for the NCBI eUtils API. 9, (2017).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Pagès, H., Aboyoun, P., Gentleman, R. & DebRoy, S. Biostrings: Efficient manipulation of biological strings. R package version 2.66.0 (2022).
Acknowledgements
This work was supported by IKERBASQUE Basque Foundation for Science (V.R.K and P.B.P) and by awards IT1657-22, (A.L-E. and V.R.K), and IT1487-22 (P.B.P.); pre-doctoral fellowships (PRE_2022_2_0105, A.L-E.; PRE_2022_2_0171, P.A.) from Consejería de Educación, Universidades e Investigación del Gobierno Vasco and Erasmus Mundus Joint Master Degree ECT+ scholarship (EMJMD 2019-1485, E.B.) from the European Union. The study was also supported by awards PID2020-118028GB-I00 (P.B.P.) and TED2021-132109B-C21 (V.R.K. and A.L-E.) from the Ministerio de Ciencia e Innovación of the Spanish Government and the European Union NextGenerationEU/PRTR as well as by BlueAdapt (ID: 101057764) European Union’s Horizon 2020 HORIZON-HLTH-2021-ENVHLTH-02-03 project.
Author information
Authors and Affiliations
Contributions
V.R.K. and P.B.P. conceived, designed, and supervised the project. A.L.-E. and E.B. compiled Vibrio data repository, treated sequences, conducted phylogenetic analyses and tested primers in silico. A.L.-E., E.B. and P.A. constructed the figures. A.L.-E. and V.R.K. conducted the experimental validation of primers. A.L.-E. led the writing and E.B. contributed to the first draft. All authors contributed to the further writing and revision of the manuscript, and approved its submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leunda-Esnaola, A., Bunin, E., Arrufat, P. et al. Harnessing the intragenomic variability of rRNA operons to improve differentiation of Vibrio species. Sci Rep 14, 9908 (2024). https://doi.org/10.1038/s41598-024-60505-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-60505-9
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.