Abstract
Organoselenium compounds are rare in nature but play important physiological roles by exploiting the distinct features of selenium. However, the ability to explore these compounds and their implications has been hindered by the limited availability of (bio)synthetic tools for the generation of organoselenium molecules, particularly the lack of enzymatic strategies for C‒Se bond formation. Here we develop an enzymatic approach for C‒Se bond formation using sulfur carrier proteins to biosynthesize the isologous selenium counterparts of cysteine, thiamine and a chuangxinmycin derivative. Our results indicate that widespread sulfur-carrier-protein-based biosynthetic systems provide promiscuous and programmable machinery for the production of unnatural Se-containing compounds. We anticipate that the ‘element engineering’ strategy used in this study will provide new opportunities to develop biologically rare molecules or abiological-element-containing chemicals not found in nature.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Experimental data supporting the conclusions of this study are available within the article and its Supplementary information. Protein sequences are retrieved from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/) with the accession numbers in Supplementary Table 1.
References
Wessjohann, L. A., Schneider, A., Abbas, M. & Brandt, W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 388, 997–1006 (2007).
Clive, D. L. J. et al. Organic tellurium and selenium chemistry. Reduction of tellurides, delenides, and delenoacetals with triphenyltin hydride. J. Am. Chem. Soc. 102, 4438–4447 (1980).
Mukherjee, A. J., Zade, S. S., Singh, H. B. & Sunoj, R. B. Organoselenium chemistry: role of intramolecular interactions. Chem. Rev. 110, 4357–4416 (2010).
Chivers, T. & Laitinen, R. S. Tellurium: a maverick among the chalcogens. Chem. Soc. Rev. 44, 1725–1739 (2015).
Weekley, C. M. & Harris, H. H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 42, 8870–8894 (2013).
Reich, H. J. & Hondal, R. J. Why nature chose selenium. ACS Chem. Biol. 11, 821–841 (2016).
Birringer, M., Pilawa, S. & Flohe, L. Trends in selenium biochemistry. Nat. Prod. Rep. 19, 693–718 (2002).
Hou, W. & Xu, H. Incorporating selenium into heterocycles and natural products—from chemical properties to pharmacological activities. J. Med. Chem. 65, 4436–4456 (2022).
Chuai, H. Y. et al. Small molecule selenium-containing compounds: recent development and therapeutic applications. Eur. J. Med. Chem. 223, 113621–113641 (2021).
Raffa, R. B. Diselenium, instead of disulfide, bonded analogs of conotoxins: novel synthesis and pharmacotherapeutic potential. Life Sci. 87, 451–456 (2010).
Tan, Y., Wang, M. & Chen, Y. Reprogramming the biosynthesis of precursor peptide to create a selenazole-containing nosiheptide analogue. ACS Synth. Biol. 11, 85–91 (2022).
Hou, W. et al. Selenium as an emerging versatile player in heterocycles and natural products modification. Drug Discov. Today 27, 2268–2277 (2022).
Jin, Z. M. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).
Amporndanai, K. et al. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun. 12, 3061–3068 (2021).
Turanov, A. A. et al. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv. Nutr. 2, 122–128 (2011).
Kayrouz, C. M., Huang, J., Hauser, N. & Seyedsayamdost, M. R. Biosynthesis of selenium-containing small molecules in diverse microorganisms. Nature 610, 199–204 (2022).
Wittwer, A. J., Tsai, L., Ching, W. M. & Stadtman, T. C. Identification and synthesis of a naturally occurring selenonucleoside in bacterial tRNAs: 5-[(methylamino)methyl]-2-selenouridine. Biochemistry 23, 4650–4655 (1984).
Schrauzer, G. N. Selenomethionine: a review of its nutritional significance, metabolism and toxicity. J. Nutr. 130, 1653–1656 (2000).
Sonawane, A. D. & Koketsu, M. Recent advances on C–Se bond-forming reactions at low and room temperature. Curr. Org. Chem. 23, 3206–3225 (2020).
Rafique, J., Canto, R. F. S., Saba, S., Barbosa, F. A. R. & Braga, A. L. Recent advances in the synthesis of biologically relevant selenium-containing 5-membered heterocycles. Curr. Org. Chem. 20, 166–188 (2016).
Beletskaya, I. P. & Ananikov, V. P. Transition-metal-catalyzed C–S, C–Se, and C–Te bond formation via cross-coupling and atom-economic addition reactions. Chem. Rev. 111, 1596–1636 (2011).
Reich, H. J., Yelm, K. E. & Wollowitz, S. Kinetics, thermodynamics, and stereochemistry of the allyl sulfoxide sulfenate and selenoxide selenenate [2,3] sigmatropic rearrangements. J. Am. Chem. Soc. 105, 2503–2504 (1983).
Collins, R. et al. Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase. PLoS ONE 7, e30581 (2012).
Johansson, A. L., Collins, R., Arner, E. S. J., Brzezinski, P. & Hogbom, M. Biochemical discrimination between selenium and sulfur 2: mechanistic investigation of the selenium specificity of human selenocysteine lyase. PLoS ONE 7, e30528 (2012).
Cheng, R. et al. Implications for an imidazol-2-yl carbene intermediate in the rhodanase-catalyzed C–S bond formation reaction of anaerobic ergothioneine biosynthesis. ACS Catal. 11, 3319–3334 (2021).
O'Leary, S. E., Jurgenson, C. T., Ealick, S. E. & Begley, T. P. O-Phospho-L-serine and the thiocarboxylated sulfur carrier protein CysO-COSH are substrates for CysM, a cysteine synthase from Mycobacterium tuberculosis. Biochemistry 47, 11606–11615 (2008).
Burns, K. E. et al. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 127, 11602–11603 (2005).
Dorrestein, P. C., Zhai, H. L., McLafferty, F. W. & Begley, T. P. The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chem. Biol. 11, 1373–1381 (2004).
Jurgenson, C. T., Begley, T. P. & Ealick, S. E. The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603 (2009).
Sasaki, E. et al. Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis. Nature 510, 427–431 (2014).
Dong, L. B. et al. Biosynthesis of thiocarboxylic acid-containing natural products. Nat. Commun. 9, 2362 (2018).
Zhang, X. et al. Biosynthesis of chuangxinmycin featuring a deubiquitinase-like sulfurtransferase. Angew. Chem. Int. Ed. 60, 24418–24423 (2021).
Jurgenson, C. T., Burns, K. E., Begley, T. P. & Ealick, S. E. Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis. Biochemistry 47, 10354–10364 (2008).
Sasaki, E. & Liu, H. W. Mechanistic studies of the biosynthesis of 2-thiosugar: evidence for the formation of an enzyme-bound 2-Ketohexose intermediate in BexX-catalyzed reaction. J. Am. Chem. Soc. 132, 15544–15546 (2010).
Klayman, D. L. & Griffin, T. S. Reaction of selenium with sodium borohydride in protic solvents. A facile method for introduction of selenium into organic molecules. J. Am. Chem. Soc. 95, 197–200 (1973).
Zhu, D. F., Zheng, W. R., Chang, H. F. & Xie, H. Y. A theoretical study on the pKa values of selenium compounds in aqueous solution. New J. Chem. 44, 8325–8336 (2020).
Cipollone, R., Ascenzi, P. & Visca, P. Common themes and variations in the rhodanese superfamily. IUBMB Life 59, 51–59 (2007).
Liu, L. P., Peng, Q. & Li, Y. D. Preparation of monodisperse Se colloid spheres and Se nanowires using Na2SeSO3 as precursor. Nano. Res. 1, 403–411 (2008).
Cupp-Vickery, J. R., Urbina, H. & Vickery, L. E. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 (2003).
Mihara, H., Kurihara, T., Yoshimura, T., Soda, K. & Esaki, N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272, 22417–22424 (1997).
Silva, I. R. et al. Formation of a ternary complex for selenocysteine biosynthesis in bacteria. J. Biol. Chem. 290, 29178–29188 (2015).
Mueller, E. G. Se-ing into selenocysteine biosynthesis. Nat. Chem. Biol. 5, 611–612 (2009).
Esaki, N. & Soda, K. Preparation of sulfur and selenium amino-acids with microbial pyridoxal-phosphate enzymes. Method. Enzymol. 143, 291–297 (1987).
Lawhorn, B. G., Mehl, R. A. & Begley, T. P. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org. Biomol. Chem. 2, 2538–2546 (2004).
Cheng, G., Bennett, E. M., Begley, T. P. & Ealick, S. E. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure 10, 225–235 (2002).
Manzetti, S., Zhang, J. & van der Spoel, D. Thiamin function, metabolism, uptake, and transport. Biochemistry 53, 821–835 (2014).
Xu, X. et al. Heterologous expression guides identification of the biosynthetic gene cluster of chuangxinmycin, an indole alkaloid antibiotic. J. Nat. Prod. 81, 1060–1064 (2018).
Fan, S. et al. Structural insights into the specific interaction between Geobacillus stearothermophilus tryptophanyl-tRNA synthetase and antimicrobial chuangxinmycin. J. Biol. Chem. 298, 101580 (2022).
Maupin-Furlow, J. A. Ubiquitin-like proteins and their roles in archaea. Trends Microbiol. 21, 31–38 (2013).
Iyer, L. M., Burroughs, A. M. & Aravind, L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 7, R60 (2006).
Tanabe, T. S., Leimkuhler, S. & Dahl, C. The functional diversity of the prokaryotic sulfur carrier protein TusA. Adv. Microb. Physiol. 75, 233–277 (2019).
Taylor, S. V. et al. Chemical and enzymatic synthesis of 1-deoxy-D-xylulose-5-phosphate. J. Org. Chem. 63, 2375–2377 (1998).
Meyer, O., Grosdemange-Billiard, C., Tritsch, D. & Rohmer, M. Synthesis and activity of two trifluorinated analogues of 1-deoxy-D-xylulose 5-phosphate. Tetrahedron Lett. 48, 711–714 (2007).
Acknowledgements
We thank J. Qu, G. Lin, J. Zhu, Z. Li and H. Sui from the State Key Laboratory of Microbial Technology at Shandong University for their guidance and help in HPLC-MS and NMR spectroscopy analyses. This work was supported by National Key Research and Development Program of China (2022YFC2804500 to X.Z.), National Natural Science Foundation of China (32025001 to S.L., 22237004 to S.L., 32000039 to X.Z.) and Shandong Provincial Natural Science Foundation (ZR2023ZD50 to X.Z., ZR2019ZD20 to S.L.).
Author information
Authors and Affiliations
Contributions
S.L. designed this study and analysed the results. X.Z. synthesized the substrates, performed the bioassays and analysed the results. F.C., J.G., S.Z. and X.W. cloned the genes, constructed the protein expression vectors and purified the proteins. X.Z. and S.L. wrote and revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Synthesis thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Thomas West, in collaboration with the Nature Synthesis team.
Extended data
Extended Data Fig. 1 Representative sulfur-containing natural products biosynthesized by SCP-involved pathways.
Additional organosulfur structures to Fig. 1.
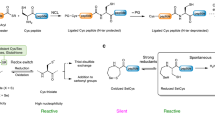
Extended Data Fig. 2 Enzymatic mechanism of the sulfur incorporation process in cysteine biosynthesis.
Key nucleophilic attack reactions are indicated by orange arrows. The sulfur source for CysO sulfuration can be either HS− (path i) or S2O32− (path ii).
Extended Data Fig. 3 Enzymatic mechanism of the sulfur incorporation process in thiamin biosynthesis.
Key nucleophilic attack reactions are indicated by orange arrows. The sulfur source for ThiS sulfuration can be either HS− (path i) or L-Cys (path ii).
Extended Data Fig. 4 Enzymatic mechanism of the sulfur incorporation process in chuangxinmycin biosynthesis.
Key nucleophilic attack reactions are indicated by orange arrows. The sulfur source for Cxm4* sulfuration can be HS− (path i), L-Cys (path ii) or S2O32− (path iii).
Extended Data Fig. 5 Sulfuration of the SCPs.
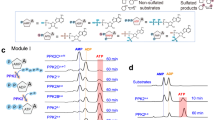
a, Schematic ATP-dependent sulfuration of CysO, ThiS, and Cxm4G (the matured form of Cxm4) by MoeZ, ThiF, and CxmM, respectively, with NaSH as the sulfur source. b, Deconvoluted HRESI-MS analysis of the sulfuration efficiency of CysO, ThiS, and Cxm4* under different pH conditions.
Extended Data Fig. 6 HRESI-MS/HPLC analysis of the recombined SCP-based selenium incorporation systems.
a, Deconvoluted HRESI-MS analyses of the selenylation efficiency of CysO (i), ThiS (ii) and Cxm4* (iii) by different activating enzymes. b, HPLC (i and iii) and HPLC-MS (ii) analysis of Se-Cys (i), Se-Thz (ii), and Se-Trp (iii) using different selenylated SCPs (that is, SCP-COSe-) as selenium donors. c, HPLC analysis of Se-Cys (i) and Se-Trp (ii) produced in the reprogrammed pathways.
Extended Data Fig. 7 Recombination in the SCP-based sulfur incorporation systems.
a, Recombination network of SCPs against different activating enzymes MoeZ, ThiF, and CxmM (i) and sulfurtransferases CysM, ThiG, and Cxm3 (ii). The left boxed histograms show the conversion ratios of SCPs (i). The right boxed histograms show the yields of organosulfur products in one-pot reactions (ii) of the three recombined pathways (the yield of Thz was not determined due to unavailability of authentic standard. ‘Positive’ indicates the product was detectable by LC-MS). The colour of each column represents the corresponding same coloured activating enzyme (i) or SCP (ii) supported reaction. b, Deconvoluted HRESI-MS analyses of the selenylation efficiency of CysO (i), ThiS (ii) and Cxm4* (iii) by different activating enzymes. c, HPLC (i and iii) and HPLC-MS (ii) analysis of the sulfur-transfer reactions of Cys (i), Thz (ii) and S-Trp (iii) by different sulfurtransferases.
Supplementary information
Supplementary Information
Supplementary Methods, Tables 1–3, Schemes 1–5, Figs. 1–41 and References.
Source data
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Cheng, F., Guo, J. et al. Enzymatic synthesis of organoselenium compounds via C‒Se bond formation mediated by sulfur carrier proteins. Nat. Synth 3, 477–487 (2024). https://doi.org/10.1038/s44160-023-00477-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44160-023-00477-2
This article is cited by
-
Making selenometabolites nature’s way
Nature Synthesis (2024)