Abstract
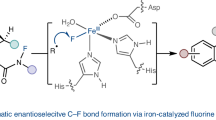
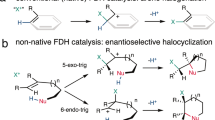
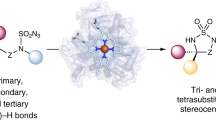
Due to the scarcity of C–F bond-forming enzymatic reactions in nature and the contrasting prevalence of organofluorine moieties in bioactive compounds, developing biocatalytic fluorination reactions represents a pre-eminent challenge in enzymology, biocatalysis and synthetic biology. Additionally, catalytic enantioselective C(sp3)–H fluorination remains a challenging problem facing synthetic chemists. Although many non-haem iron halogenases have been discovered to promote C(sp3)–H halogenation reactions, efforts to convert these iron halogenases to fluorinases have remained unsuccessful. Here we report the development of an enantioselective C(sp3)–H fluorination reaction, catalysed by a plant-derived non-haem enzyme 1-aminocyclopropane-1-carboxylic acid oxidase (ACCO), which is repurposed for radical rebound fluorination. Directed evolution afforded a C(sp3)–H fluorinating enzyme ACCOCHF displaying 200-fold higher activity, substantially improved chemoselectivity and excellent enantioselectivity, converting a range of substrates into enantioenriched organofluorine products. Notably, almost all the beneficial mutations were found to be distal to the iron centre, underscoring the importance of substrate tunnel engineering in non-haem iron biocatalysis. Computational studies reveal that the radical rebound step with the Fe(III)–F intermediate has a low activation barrier of 3.4 kcal mol−1 and is kinetically facile.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the main text and Supplementary Information. Plasmids encoding evolved ACCO variants reported in this study are available for research purposes from Y.Y. under a material transfer agreement with the University of California, Santa Barbara. The solid-state structure of 2a is available free of charge from the Cambridge Crystallographic Data Centre under reference number CCDC 2294341. All the protein structures used are available from the Protein Data Bank using their accession numbers.
References
Renata, H., Wang, Z. J. & Arnold, F. H. Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chem. Int. Ed. 54, 3351–3367 (2015).
Brandenberg, O. F., Fasan, R. & Arnold, F. H. Exploiting and engineering hemoproteins for abiological carbene and nitrene transfer reactions. Curr. Opin. Biotechnol. 47, 102–111 (2017).
Leveson-Gower, R. B., Mayer, C. & Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. 3, 687–705 (2019).
Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal. 3, 203–213 (2020).
Klaus, C. & Hammer, S. C. New catalytic reactions by enzyme engineering. Trends Chem. 4, 363–366 (2022).
Zhou, Q., Chin, M., Fu, Y., Liu, P. & Yang, Y. Stereodivergent atom-transfer radical cyclization by engineered cytochromes P450. Science 374, 1612–P1616 (2021).
Fu, Y. et al. Engineered P450 atom-transfer radical cyclases are bifunctional biocatalysts: reaction mechanism and origin of enantioselectivity. J. Am. Chem. Soc. 144, 13344–13355 (2022).
Fu, W. et al. Enzyme-controlled stereoselective radical cyclization to arenes enabled by metalloredox biocatalysis. Nat. Catal. 6, 628–636 (2023).
Rui, J. et al. Directed evolution of nonheme iron enzymes to access abiological radical-relay C(sp3)–H azidation. Science 376, 869–874 (2022).
Ren, X. & Fasan, R. Engineered and artificial metalloenzymes for selective C–H functionalization. Curr. Opin. Green Sustain. Chem. 31, 100494 (2021).
Emmanuel, M. A. et al. Photobiocatalytic strategies for organic synthesis. Chem. Rev. 123, 5459–5520 (2023).
Harrison, W., Huang, X. & Zhao, H. Photobiocatalysis for abiological transformations. Acc. Chem. Res. 55, 1087–1096 (2022).
Cheng, L. et al. Stereoselective amino acid synthesis by synergistic photoredox-pyridoxal radical biocatalysis. Science 381, 444–451 (2023).
Wang, J. et al. Fluorine in pharmaceutical industry: fluorine-containing drugs introduced to the market in the last decade (2001–2011). Chem. Rev. 114, 2432–2506 (2014).
Gillis, E. P., Eastman, K. J., Hill, M. D., Donnelly, D. J. & Meanwell, N. A. Applications of fluorine in medicinal chemistry. J. Med. Chem. 58, 8315–8359 (2015).
Zhou, Y. et al. Next generation of fluorine-containing pharmaceuticals, compounds currently in phase II–III clinical trials of major pharmaceutical companies: new structural trends and therapeutic areas. Chem. Rev. 116, 422–518 (2016).
Ogawa, Y., Tokunaga, E., Kobayashi, O., Hirai, K. & Shibata, N. Current contributions of organofluorine compounds to the agrochemical industry. iScience. 23, 101467 (2020).
Inoue, M., Sumii, Y. & Shibata, N. Contribution of organofluorine compounds to pharmaceuticals. ACS Omega 5, 10633–10640 (2020).
Wu, L., Maglangit, F. & Deng, H. Fluorine biocatalysis. Curr. Opin. Chem. Biol. 55, 119–126 (2020).
Walker, M. C. & Chang, M. C. Y. Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev. 43, 6527–6536 (2014).
Walker, M. et al. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science 341, 1089–1094 (2013).
Sirirungruang, S. et al. Engineering site-selective incorporation of fluorine into polyketides. Nat. Chem. Biol. 18, 886–893 (2022).
Rittner, A. et al. Chemoenzymatic synthesis of fluorinated polyketides. Nat. Chem. 14, 1000–1006 (2022).
Szpera, R., Moseley, D. F. J., Smith, L. B., Sterling, A. J. & Gouverneur, V. The fluorination of C–H bonds: developments and perspectives. Angew. Chem. Int. Ed. 58, 14824–14848 (2019).
Leibler, I. N.-M., Gandhi, S. S., Tekle-Smith, M. A. & Doyle, A. G. Strategies for nucleophilic C(sp3)–(radio)fluorination. J. Am. Chem. Soc. 145, 9928–9950 (2023).
Huang, X. et al. Late stage benzylic C–H fluorination with [18F]fluoride for PET imaging. J. Am. Chem. Soc. 136, 6842–6845 (2014).
Park, H., Verma, P., Hong, K. & Yu, J.-Q. Controlling Pd(IV) reductive elimination pathways enables Pd(II)-catalysed enantioselective C(sp3)–H fluorination. Nat. Chem. 10, 755–762 (2018).
Britton, R. et al. Contemporary synthetic strategies in organofluorine chemistry. Nat. Rev. Methods Primers 1, 47 (2021).
O’Hagan, D. & Young, R. J. Future challenges and opportunities with fluorine in drugs? Med. Chem. Res. 32, 1231–1234 (2023).
O’Hagan, D. & Deng, H. Enzymatic fluorination and biotechnological developments of the fluorinase. Chem. Rev. 115, 634–649 (2015).
Vaillancourt, F. H., Yeh, E., Vosburg, D. A., Garneau-Tsodikova, S. & Walsh, C. T. Nature’s inventory of halogenation catalysts: oxidative strategies predominate. Chem. Rev. 106, 3364–3378 (2006).
Agarwal, V. et al. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 117, 5619–5674 (2017).
Crowe, C. et al. Halogenases: a palette of emerging opportunities for synthetic biology–synthetic chemistry and C–H functionalisation. Chem. Soc. Rev. 50, 9443–9481 (2021).
Papadopoulou, A., Meyer, F. & Buller, R. M. Engineering Fe(II)/α-ketoglutarate-dependent halogenases and desaturases. Biochemistry 62, 229–240 (2023).
Hayashi, T. et al. Evolved aliphatic halogenases enable regiocomplementary C–H functionalization of a pharmaceutically relevant compound. Angew. Chem. Int. Ed. 58, 18535–18539 (2019).
Vaillancourt, F. H., Vosburg, D. A. & Walsh, C. T. Dichlorination and bromination of a threonyl-S-carrier protein by the non-heme FeII halogenase SyrB2. ChemBioChem 7, 748–752 (2006).
Matthews, M. L. et al. Direct nitration and azidation of aliphatic carbons by an iron-dependent halogenase. Nat. Chem. Biol. 10, 209–215 (2014).
Gomez, C. A., Mondal, D., Du, Q., Chan, N. & Lewis, J. C. Directed evolution of an iron(II)- and α-ketoglutarate-dependent dioxygenase for site-selective azidation of unactivated aliphatic C–H bonds.Angew. Chem. Int. Ed. 62, e202301370 (2023).
Chan, N. H. et al. Non-native anionic ligand binding and reactivity in engineered variants of the Fe(II)- and α-ketoglutarate-dependent oxygenase, SadA. Inorg. Chem. 61, 14477–14485 (2022).
Zhan, C.-G. & Dixon, D. A. Hydration of the fluoride anion: structures and absolute hydration free energy from first-principles electronic structure calculations. J. Phys. Chem. A 108, 2020–2029 (2004).
Groendyke, B. J., AbuSalim, D. I. & Cook, S. P. Iron-catalyzed, fluoroamide-directed C–H fluorination. J. Am. Chem. Soc. 138, 12771–12774 (2016).
Pinter, E. N., Bingham, J. E., AbuSalim, D. I. & Cook, S. P. N-directed fluorination of unactivated Csp3–H bonds. Chem. Sci. 11, 1102–1106 (2020).
Ge, W. et al. Isopenicillin N synthase mediates thiolate oxidation to sulfenate in a depsipeptide substrate analogue: implications for oxygen binding and a link to nitrile hydratase? J. Am. Chem. Soc. 130, 10096–10102 (2008).
Wilmouth, R. C. et al. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10, 93–103 (2002).
Gopal, B., Madan, L. L., Betz, S. F. & Kossiakoff, A. A. The crystal structure of a quercetin 2,3-dioxygenase from Bacillus subtilis suggests modulation of enzyme activity by a change in the metal ion at the active site(s). Biochemistry 44, 193–201 (2005).
Zhang, Z., Ren, J.-S., Clifton, I. J. & Schofield, C. J. Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase—the ethylene-forming enzyme. Chem. Biol. 11, 1383–1394 (2004).
Houben, M. & Van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 10, 695 (2019).
Vaillancourt, F. H., Yin, J. & Walsh, C. T. SyrB2 in syringomycin E biosynthesis is a nonheme FeII α-ketoglutarate- and O2-dependent halogenase. Proc. Natl Acad. Sci. USA 102, 10111–10116 (2005).
Blasiak, L. C., Vaillancourt, F. H., Walsh, C. T. & Drennan, C. L. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature 440, 368–371 (2006).
Mitchell, A. J. et al. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat. Chem. Biol. 12, 636–640 (2016).
Liu, X. in Methods in Enzymology, Vol. 604 (ed. Moore, B. S.) 389–404 (Academic Press, 2018).
Voss, M. et al. Enzyme engineering enables inversion of substrate stereopreference of the halogenase WelO5*. ChemCatChem 14, e202201115 (2022).
Büchler, J. et al. Algorithm-aided engineering of aliphatic halogenase WelO5* for the asymmetric late-stage functionalization of soraphens. Nat. Comm. 13, 371 (2022).
Duewel, S. et al. Directed evolution of an FeII-dependent halogenase for asymmetric C(sp3)–H chlorination. ACS Catal. 10, 1272–1277 (2020).
Hillwig, M. L. & Liu, X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol. 10, 921–923 (2014).
Marchand, J. A. et al. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–424 (2019).
Neugebauer, M. E. et al. A family of radical halogenases for the engineering of amino-acid-based products. Nat. Chem. Biol. 15, 1009–1016 (2019).
Neugebauer, M. E. et al. Reaction pathway engineering converts a radical hydroxylase into a halogenase. Nat. Chem. Biol. 18, 171–179 (2022).
Knorrscheidt, A. et al. Accessing chemo- and regioselective benzylic and aromatic oxidations by protein engineering of an unspecific peroxygenase. ACS Catal. 11, 7327–7338 (2021).
Usharani, D., Janardanan, D. & Shaik, S. Does the TauD enzyme always hydroxylate alkanes, while an analogous synthetic non-heme reagent always desaturates them? J. Am. Chem. Soc. 133, 176–179 (2011).
Lanzalaco, S. et al. Atom transfer radical polymerization with different halides (F, Cl, Br, and I): is the process ‘living’ in the presence of fluorinated initiators? Macromolecules 50, 192–202 (2017).
Aoto, Y. A., de Lima Batista, A. P., Kohn, A. & de Oliveira-Filho, A. G. S. How to arrive at accurate benchmark values for transition metal compounds: computation or experiment? J. Chem. Theory Comput. 13, 5291–5316 (2017).
Acknowledgements
This research is supported by the NIH (R35GM147387 to Y.Y.), NSF (CHE-2247505 to P.L.) and Boehringer Ingelheim’s IU More Green Grant (BI number 763955 to Y.Y.). We acknowledge the NSF BioPACIFIC MIP (DMR-1933487) and NSF MRSEC at the University of California, Santa Barbara (DMR-2308708) for access to instrumentation. Computational studies were carried out at the University of Pittsburgh Center for Research Computing and the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, supported by NSF award numbers OAC-2117681, OAC-1928147 and OAC-1928224. We thank Y. Wang (University of Pittsburgh) for critical reading of this paper.
Author information
Authors and Affiliations
Contributions
Y.Y. conceived and directed the project. L.-P.Z. performed all the enzyme engineering, Michaelis–Menten kinetics, substrate synthesis and substrate scope studies. Y.Y., L.C. and Y. Zhao performed enzyme mining. L.-P.Z. and L.C. performed initial enzyme evaluation. F.G. and R.G. provided some substrates. B.K.M. carried out the computational studies with P.L. providing guidance. H.W. and Y. Zhang participated in discussions and provided suggestions. Y.Y., L.-P.Z., P.L. and B.K.M. wrote the paper with the input of all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Synthesis thanks Kyle Biegasiewicz, Hans Senn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Thomas West, in collaboration with the Nature Synthesis team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Tables 1–16, experimental details, X-ray crystallographic analysis details, computational details, NMR spectra and HPLC analysis.
Supplementary Data 1
Crystallographic data for compound 2a, CCDC 2294341.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, LP., Mai, B.K., Cheng, L. et al. Biocatalytic enantioselective C(sp3)–H fluorination enabled by directed evolution of non-haem iron enzymes. Nat. Synth (2024). https://doi.org/10.1038/s44160-024-00536-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44160-024-00536-2