Abstract
The use of novel sequencing and high-throughput techniques has become widespread, and are now readily available to obtain the comprehensive transcription profile of the human genome. Noncoding RNAs (ncRNAs) are transcripts that have no apparent protein-coding capacity, but they have important roles in human physiology. Most research in this area has focused on micro-RNAs. However, the role of long ncRNAs (lncRNAs) as drivers of tumor suppression and oncogenic functions has recently been examined in numerous cancer types. Epigenetic alterations can reportedly deregulate the expression of any type of transcript. However, the exact mechanisms of epigenetic regulation of lncRNA are still unknown. In this review, the authors primarily focus on the epigenetic effects modulating ncRNA in colorectal cancer (CRC). The authors specifically discuss examples of oncogenic ncRNA in CRC pathobiology, as well as its extended diagnosis, prognosis and therapy.
Similar content being viewed by others
Introduction
The best-studied sequences in the human genome have generally been protein-coding genes that correspond to only 1.5–2.0% of the entire genome.1 Recently, non-protein-coding regions have emerged as new areas of study, and it is now clear that these regions of the genome have essential functions in normal development and in diseases, including cancer. Most of the sequences in the transcriptome are noncoding RNAs (ncRNAs) that are not translated into proteins. They can be classified as either small ncRNAs including most microRNAs (miRNAs) or as long ncRNA (lncRNA). The latter comprises a very heterogeneous group of ncRNAs that are more than 200 nt in length. They include, among others, long intergenic ncRNAs (lincRNAs), transcribed ultraconserved regions (T-UCRs), pseudogenes and antisense RNAs.2
Expression of ncRNA is regulated by epigenetic changes that consist of DNA methylation and histone modifications. DNA methylation represses gene expression in cancer cells. Methylation, which is reversible, occurs on cytosines at carbon position 5. Methylation is carried out by three DNA methyltransferases (DNMT1, DNMT3A or DNMT3B). Methylation of promoter-associated CpG islands usually reduces transcription levels of the corresponding gene in cancer cells.3 Histone proteins are the main constituents of chromatin and undergo posttranslational modifications. This modification determines and allows access to chromatin. The early replicating form, called euchromatin, is accessible, and the late replicating form, called heterochromatin, is inaccessible. These histone modifications constitute the ‘histone code’. For instance, euchromatin accompanies acetylation of histones H3 and H4 and methylation of lysine 4 of histone H3 (H3K4me), whereas di- or trimethylation of lysine 9 of histone 3 (H3K9me) occurs in heterochromatin. Moreover, several kinds of histone deacetylases (HDACs) and histone methyltransferases influence and control these histone modifications. Eventually, trimethylating histone H3 lysine 27(H3K27me3) regulates the silencing of the epigenetic gene in conjunction with the polycomb repressive complex 2 (PRC2).4
Alterations in the epigenetic regulation of ncRNA are pivotal in the pathogenesis of human disease.5 This is not surprising as DNA is globally hypomethylated in cancer and there are alterations in the epigenetic regulation of ncRNAs in the pathogenesis of cancer.5 Both miRNAs and long ncRNAs are modulated by hypermethylation6, 7 or hypomethylation.8 Furthermore, many lncRNAs interact with histone-modifying and chromatin-remodeling complexes, and miRNAs can target genes that are important in the epigenetic machinery.9
Although Vogelstein’s model has been accepted worldwide as an important paradigm whose sequence of genetic change leads to colorectal cancer (CRC), and despite the progress being made in basic and clinical research and the numerous published reports, the roles and mechanisms of action of ncRNAs in the pathogenesis of CRC are still unclear. In particular, lncRNAs are poorly characterized, and it is still a key challenge to understand their precise roles in relation to CRC biological characteristics. In this review, despite being just the tip of the iceberg, we focus on epigenetics and the roles of miRNAs and lncRNAs in the tumorigenesis of CRC, including possible diagnostic and/or prognostic factors. Moreover, we summarize new prospects for miRNAs and long ncRNAs and describe mechanisms that cause abnormal expression of ncRNAs in CRC.
miRNAs and cancer
The most widely studied and well-known classes of ncRNAs are miRNAs, which are small ncRNAs that are only ~22 nt in length. They function through gene silencing after transcription by modulating mRNA and translating it into proteins.10 miRNAs control the translation of over 60% of protein-coding genes. Some miRNAs control the gene expression to bind specific individual targets; moreover, others can operate as central regulators of a gene expression procedure. For instance, specific miRNAs control the expression levels of hundreds of genes simultaneously, cooperatively and comprehensively.10 The first relation between cancer and ncRNAs was investigated by Calin et al. in 2002. They disclosed the genome region in which transcripts miR-15 and miR-16 were found to be frequently absent in chronic lymphocytic leukemia (CLL).11 Right after that study, the first miRNA microarray emerged,12 and it successfully revealed miRNA profiling in miRNA. These profiles were able to distinguish tumors from normal counterparts.13, 14, 15
Calin et al. used comprehensive microarray profiling using 94 samples comprising 94 patients with CLL and constituted the interesting gene expression panel of nine miRNAs. These expression patterns, which vary and depend on different phases and breakpoints of disease progression in CLL, may implicate the timing of the beginning of treatment.16
Recently, lliou et al. found that DICER, which is necessary for the growth and maturation of miRNA, has an important role in stem cell properties. Attenuation of DICER1 in colon cancer cell lines increased the stem cell markers for colon cancer and decreased several specific miRNAs. Both in vitro and in vivo studies disclose that knockdown of DICER1 induces carcinogenesis and metastatic ability without proliferation itself, which strongly implicates its role in cancer stem cells.17
Epigenetics and miRNA in CRC
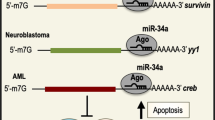
To clarify the mechanisms that underlie the aberrant miRNA expression in malignancy, numerous studies have demonstrated the regulation system of miRNA. Currently, it is reported that miRNAs are subject to the same epigenetic regulatory mechanisms as conventional protein-coding genes. Moreover, aberrant epigenetic regulation—for example, CpG island hypermethylation—affects abnormal miRNA expression in cancers (Figure 1a). Moreover, a subgroup of these miRNAs have an effect on the expression of epigenetic effectors—for instance, DNMTs, HDACs and polycomb genes—and they are named epi-miRNAs (Figure 1b).
Fabbri et al. first investigated the existence of epi-miRNAs in lung cancer cell lines. They reported that miR-29a, -29b and -29c directly targeted and controlled the epigenetic regulator.18 Re-expression of miR-29 disrupted DNA methylation and led to general hypomethylation status. Moreover, in an AML model, Garzon et al.19 reported that miR-29b directly targeted SP1, which is one of the trans-activators of the DNMT1 gene, and indirectly silenced DNMT.
miR-34
A member of the miR 34 family, miR 34a resides in the second exon of EF570049, which has a p53-binding site and CpG island in the promoter region. Despite DNA methylation of the EF570049 promoter CpG island in CLL, miR-34a was upregulated. Importantly, loss of methylation and enrichment of H3K4me3 were observed in a region 12 Kb upstream of the miR-34a coding sequence.20
The miR-34 family has been attracting attention because of the substantial correlation and cross-talk with p53.9, 21 Indeed, dysfunction of p53 influences and induces depressed miR-34a, the cross-talk of which was observed in many types of human diseases including CRC.22, 23 miR-34a can affect CRC invasion and metastasis in conjunction with IL6R, ZNF281, MET, snail family zinc finger 1 and 2 (SNAI1, SNAI2) and β-catenin (CTNNB1).24, 25, 26, 27
Lujambio et al. used three frequent aggressive cell lines under treatment with 5-AZA and identified cancer-specific CpG island hypermethylation of the promoter lesion with three transcribed miRNAs: miR-148a, miR-34b/c and miR-9. The rescue experiment of the miR-34b/c cluster in cancer cell lines with epigenetic depression reduced the cell motility and metastatic frequency in an in vitro study.28 miR 34b/c resides on chromosome 11 as a dicistronic cluster within the transcription unit BC021736, and only the expression of miR-34b/c was affected by pharmacologic demethylation. Moreover, the miR-34b/c cluster is epigenetically regulated in the colon cancer cell line HCT116.29 Indeed, Siemens et al. showed that, in CRC, the presence and frequency of metastasis in clinical patients is significantly correlated with attenuated miR-34a expression, which was induced by abounding CpG methylation of the promoter lesion of miR- 34a and miR-34b/c.26
miR-200
The miR-200 family consists of five submembers of miRNAs, miR-200a, miR-200b, miR-200c, miR-429 and miR-141, which are transcripted from two different genome locations, chromosome 1 and chromosome 12.30, 31 These members maintain an epithelial phenotype and deregulate the epithelial-to-mesenchymal transition (EMT). The attenuation of miRNA is associated with mesenchymal features as observed in EMT and tumor progression, including an aggressive cancer phenotype.32 In breast and prostate cancer cell lines, the observed abnormal DNA methylation of the miR-200c and miR-141 at CpG island is strongly associated with their inadequate silencing. Moreover, epigenetic regulation of this miRNA cluster is conserved evolutionarily according to mouse experiments.33 In lung cancer cell lines, there is no variation with miRNA expression under treatment with demethylating agents or HDAC inhibitors.34
In CRC, miR-200c also has a pivotal role (that is, EMT phenomenon) with respect to cancer aggressive ability, including cell proliferation, invasion, migration and metastasis. Hur et al. reported that the miR-200c expression level of the aggressive CRC clinical tissue and high frequent metastasis CRC cell lines is attenuated significantly. Overexpression study using transfection techniques with precursors of miR-200c to RKO and SW620 showed that miR-200c accelerated the proliferation of CRC cancer cells but did not affect the ability of invasion and migration. Moreover, miR-200c targets zinc finger E-box binding homeobox 1/2 (ZEB1/2), which is one of the EMT-associated molecules. In CRC cell lines and clinical CRC including liver metastasis, this reduction induced the attenuated EMT phenomenon by E-cadherin overexpression and reduced vimentin expression.35 In stem cell biology, miR-200c is well known as the target of SOX2, which is essential for differentiation of embryonic stem cells. Attenuation of miR-200c in the cancer cell line induced sphere formation, which is a typical morphological change in stem cells and in other stem cell-associated molecules including SOX2 via the P13K/AKT pathway.36 According to the above reports, CRC patients with higher miR-200c expression had significantly worse survivals compared with patients with lower expression.37
Let-7
The let-7 family has been well known as the most popular conserved miRNA.38 It has also been reported to have tumor-suppressive effects in various malignancies.39, 40 The let-7 family was found for the first time to be essential for the development in Caenorhabditis elegans. Although developed let-7 has been highly conserved until now, the members of this family undergo changes. For instance, the human let-7 family has 10 mature miRNAs produced from 13 precursor sequences.41 Whereas the expression of let-7a-3 is kept low by canonical silencing by promoter hypermethylation in normal cell lines, the expression is released in the colon cancer cell line HCT-116 without DNMT1 and DNMT3B.8 In ovarian cancer, Lu et al.42 disclosed that hypermethylation of the promoter lesion of the let-7a-3 was frequent and that it had a positive relation with the expression of insulin-like growth factor-II and with prognosis.
The relationship between the let-7 family and RAS, which is one of the most famous oncogenes, including in CRC, has been disclosed.41, 43, 44 KRAS (representative RAS) has been found to be a small monomeric GTPase and controls signal transduction with respect to proliferation.45 RAS mutations and amplifications are also frequently found in CRC patients,46 and it has an important role in the sequential step toward colorectal carcinogenesis advocated by Vogelstein.47 In the last decade, evaluation of the KRAS mutation status was considered the gold standard to determine the presence of the anti-EGFR antibody (that is, Cetuximab and Panitumab) in CRC.48 Actually, Regusa et al.49 reported inhibited expression of let-7b and let-7e in the Cetuximab-resistant CRC cell line and under Cetuximab treatment. In later years, the polymorphism in the 3′-UTR of KRAS mRNA implied its capacity to predict outcome from CRC. Thus far, the results have been controversial and conflicting.44, 50, 51, 52 Han disclosed that the expression of let-7c conversely associated with TMN stage, tumor metastasis and poor prognosis in primary CRC tissue, and let-7c targets KRAS, MMP1 and PBX3 directly on the luciferase reporter assay. Moreover, the ectopic and rescue experiment also confirmed the tumor-suppressive function of let-7c, which inhibits metastasis in vivo and in vitro.53 Comprehensive gene expression analysis of miRNA by Svoboda et al has uncovered the impact of let-7e with regard to the sensitivity and resistance to neoadjuvant therapy.54 Overexpression of let-7e has implications in it being a responder to CRC. Moreover, Ogata-Kawata et al. evaluated the serum levels of some microRNAs to determine which of these mRNAs are appropriated diagnostic tools for CRC. They identified the miRNA group that was emitted in CRC patients' serum samples compared with healthy volunteers.55 Taken together, the let-7 family has the potential to elucidate CRC oncogenesis and pathogenesis, which can lead to the development of new noninvasive diagnostic tools in the near future.
miR-1 (epi-miRNA)
Epi-miRNAs described in the earlier paragraph are generally involved in modulating the expression of HDACs and PRC genes. For instance, both miR-1 and miR-140 directly bind to the mRNA of HDAC4.56, 57 Moreover, Migliore et al. found that the expression of miR-1 is attenuated in 84% of CRC tissue compared with that in corresponding normal tissues and the expression level strongly correlates with MET, which is overexpressed tyrosine kinase receptor for hepatocyte growth factor, and also well known to lead to CRC aggressiveness. Moreover, in vitro ectopic and knockdown study showed the anti-oncogenic effect of miR-1 from assay for viability, migration and invasion caused by MET.58
miR-101 (epi-miRNA)
EZH2 has the catalytic ability for PRC2 and accounts for heterochromatin modification by trimethylating histone H3 lysine 27 (H3K27me3). Varambally et al. reported that the expression of miR-101 is inhibited during cancer progression in cancer cell lines and primary tumors related to prostate cancer and correlates with the augmentation of EZH2 gene expression. These findings implicated miR-101 as an epi-miRNA. This hypothesis was confirmed by experiments in vitro and in vivo, which found that miR-101 directly targets EZH2.59, 60 In CRC, miR-101 also functions as a tumor suppressor in both cell lines and cancer tissue.61, 62 miR-101 targets and negatively regulates prostaglandin E receptor 4 (EP4) and COX2. In colon cancer and normal tissue, the expression level of miR-101 and EP4 has inverse relation. miR-101 induced the reduction of cancer cell proliferation and motility in ectopic and knockdown assay.63 Strillacci et al. implied the inactivated potency for CRC progression with miR-101. They disclosed that miR-101 exerts its effect on β-catenin amassment in the nucleus, and then inhibits cell proliferation, invasion ability and survival at hypoxic conditions in conjunction with promotion of cell adhesion-related E-cadherin and ZEB1 expression. Taken together, this implies that miR-101 has important roles in EMT.62
miR-148 (epi-miRNA)
DNMT3b expression is regulated by the control of miR-148a and miR-148b. The occupying by miR-148 in the gene-coding region of mRNA lead to transcribe several splicing variants from.64 Curiously, the promoter region of miR-148a frequently undergoes hypermethylation as well as epigenetic regulation in several cancers.28 In CRC, miR-148a upregulation induces apoptosis through BCL2 inhibition.65 Conversely, miR-148a downregulation in CRC is associated with increased tumor size.66
Epigenetic regulation of lncRNA in CRC
LncRNAs and their functions are under intense investigation.2 However, the actual mechanism has not been identified because of the heterogeneity of lncRNAs. LncRNAs were originally recognized as RNA molecules longer than 200 nt without coding a protein. However, this cutoff was arbitrarily chosen and was not based on functionality. Besides, this understanding might be sometimes too simple and does not consider RNA purification protocols and the overlap between lncRNAs and open reading frames.67
A recent review68 described the myriad functions of lncRNAs. Four types of molecular mechanisms were distinguished: signals, decoys, guides and scaffolds. Signals and decoys include those working as molecular blocks or sponges to pull away RNA-binding proteins, including transcription factors, chromatin modifiers or other regulatory proteins. Guides recruit chromatin-modifying enzymes to target genes, either in cis near the site of lncRNA production or in trans to distant target genes and scaffolds.
Various mechanisms have been proposed to account for the transcriptional regulation of gene expression by lncRNAs. LncRNAs are widely accepted as mediating epigenetic modifications of DNA by changing the chromatin status and replicating form.69 At the human HOX loci, hundreds of lncRNAs exert and cooperate with RNA polymerase and histone modification enzymes to modulate the chromatin status.70 X-chromosome inactivation is generally and widely well known as one of the physiological processes in mammals; the X-inactivation-specific transcript (Xist) lncRNA was investigated. The Xist is transcribed from the X-chromosome itself and encourages the polycomb complex to silence the X chromosome.71 Interestingly, Yildirim et al. found in mice that loss of Xist resulted in X reactivation and consequent genome-wide changes. Moreover, deleting Xist induced aberrant maturation of hematopoietic stem cells and extreme neoplasm in myelo-proliferation and myelodysplastic syndrome.72
In addition, we should not confuse lncRNAs with lincRNAs,73 which are categorized as a subtype of lncRNA and transcribed from intergenic regions. LincRNAs were initially described under control of histone mark signatures, specifically trimethylation in lysine 4 (H3K4m3) and lysine 36 of histone 3 (H3K36m3 and K4K36). Nearly 3000 uncovered lincRNAs have been reported to exist in mouse and human cell lines in the first reports about LincRNAs.73, 74 However, many unknown lincRNAs might exist and should be disclosed in other backgrounds.75
About 20% of lincRNAs bind to PRC2 and control the gene expression in conjunction with several polycomb proteins toward DNA regions by varying the histone and chromatin structure and suppressing transcription activity.74 Recent reports suggest that lincRNAs bind directly to the polycomb proteins and constitute PRCs, and then influence epigenetic silencing with the DNA-specific region (Figure 2a). However, whether the lincRNA-polycomb complex is aware of the target DNA position is still a matter of debate.76 Besides, it is still unknown how and which transcription factors bind lincRNAs and the relationship between RNA-binding proteins and lincRNAs as they conduct with other noncoding RNAs.77
Examples of long intergenic RNA function. (a) Long intergenic RNAs (lincRNAs) transcribed from intergenic regions can coordinate and recruit epigenetic or histone-modifying complexes, including polycomb repressive complexes (PRCs), via transrepression. The bottom of the panel indicates the peak diagram for CHIP-sequence experiments identifying histone modifications: H3K4me3, trimethylation at lysine 4 of histone 3 (found near promoters); H3K36me3, trimethylation at lysine 36 of histone 3 (found near-active transcripts). (b) Transcription of antisense-noncoding RNAs (AS-ncRNAs) from protein-coding genes (PCGs) but in the opposite direction. This appears to regulate gene expression by epigenetic change via cis regulation.
Hox transcript antisense intergenic RNA
HOTAIR (Hox transcript antisense intergenic RNA) is one of the most popular lincRNAs located and transcribed from the homeobox C gene cluster on chromosome 12, and its biological function in human malignancy has been revealed among that of recent noncoding RNAs.78 The overexpression of HOTAIR induces polycomb protein and targets at the genome region in epithelial cancer cells.79 The capacity for invasion and metastasis is also enhanced in these cells, and both are subject to PRC2. Conversely, cancer invasiveness is decreased without HOTAIR expression.80 Overall, HOTAIR may have an important function in regulating epigenetics and in mediating cell transformation in malignancy. CRC patients with overexpression of HOTAIR had a poor prognosis compared with patients with a low expression. Kogo et al.81 showed that the overexpression group among CRC patients exhibited poorer pathological differentiation of tumor, greater tumor sizes, more frequent liver metastasis and worse prognosis compared with the attenuated expression group.
Colorectal neoplasia differentially expressed
Similar to HOTAIR, CRNDE (colorectal neoplasia differentially expressed) transcribed from chromosome 16 and into multiple transcript variants. Moreover, it has epigenetic capacity with PRC2. In the early phase of mammalian development, CRNDE expression levels rise, and decrease gradually after that. Therefore, it is necessary to maintain pluripotency in mouse embryonic stem cells when associated with CRC pathogenesis.75 CRNDE-h, one of the several splice variants of CRNDE, is a feasible diagnostic tool with sufficient sensitivity and specificity for distinguishing adenomas or carcinomas from normal tissues.82
Metastasis-associated lung adenocarcinoma transcript 1
ONE of the well-known lncRNAs related to malignancy is MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) on chromosome 11.83 It appears and functions in the nucleus and has an effect on pre-mRNA metabolism associated with SC35 splicing domains.84 Moreover, MALAT1 has an important role in tumor suppressor proteins.85 In addition, it has also been found to be a regulator of E2F1, which is a crucial transcriptional factor in modulating cell cycling and tumor-suppressor proteins.86 In vitro studies showed that attenuation of MALAT1 affected the rate of bladder cancer cell migration along with EMT-associated molecules—for instance, ZEB1, ZEB2 and SNAILl. Moreover, depression of MALT1 induces cell death at the G2/M phase with aberrant mitosis.87 Recently, Wilisz et al. investigated and named MALAT1-associated small cytoplasmic RNA (mascRNA) as the primary transcription product in a 3′-end processing mechanism of the MALAT1 at a 6.7-kb nuclear-retained lncRNA and a cytoplasmic 61-nt tRNA-like ncRNA.88 Point mutations of MALAT1 and MALAT1 RNA fragment containing mascRNA were observed with high frequency in CRC cell lines and cancer tissues. Interestingly, the abounding MALAT1 RNA fragment in CRC cells promotes cell proliferation and invasion.89 Moreover, in CRC patients, Zheng et al. reported that overexpression of MALAT1 was asscociated with distant metastasis and worse prognosis compared with low expression.90
Antisense ncRNA
Morris and Vogt reviewed another class of lncRNAs that includes antisense transcripts and regulates gene expression by changes in chromatin status.91 Antisense ncRNA transcripts from overlap protein-coding gene in the opposite direction. Although it is widely accepted that small-interfering RNA functions to degrade messenger RNA generally, recent antisense ncRNA also seems to have an important role as it modulates the epigenetic change at the promoter site of the sense transcript (Figure 2b). The existence of antisense transcription with epigenetic gene silencing has been investigated in several oncogenes or tumor-suppressor genes; for example, p21, c-Myc, p15, p53, TIE1 and PU.1 have antisense transcription and consequent transcription gene silencing.92
As an example, ANRIL from the INK4A/ARF tumor-suppressor locus on chromosome 9 was initially described from the genome region where it overserved the deletion in hereditary neural system tumors.93 ANRIL was also recognized as a polyadenylated lncRNA antisense to the CDKN2A and CDKN2B genes. On the other hand, two groups found that ANRIL binds to CBX7 and SUZ12 directly and represses the INK4A/INK4B (p15) isoforms by repression of histone modifications in vivo.94, 95 However, these results were generated from different cell types, and it is still unknown as to whether ANRIL binds both complexes simultaneously. Future studies on CRC pathogenesis, diagnosis and patient prognosis are warranted.
Conclusion
This review article primarily focused on and interpreted recent studies on ncRNAs, emphasizing epigenetic regulation and CRC. Biological understanding of ncRNAs initially proceeded slowly, but there is increasing recognition that ncRNAs modulate the expression of genes critical to the oncogenic process. In cancer, ncRNA expression regulates epigenetic changes and is therefore involved in the entire spectrum of disease. In particular, aberrant lncRNA function disrupts ordinary cell biology by encouraging epigenetic depressions of downstream target genes. The discovery of ncRNA with high-throughput technologies has furthered the current understanding of transcriptome complexity. Curiously, The ENCODE consortium constituted the profiling of a variety of cell lines for 12 histone modifications and variants including H3K4me3 and acetylation of histone 3 at lysine 9 (H3K9ac) to better understand regulatory regions in the human genome.96 Integrating data profiles from different sources will allow researchers to estimate the global influence of epigenetics on the regulation of ncRNAs and aberrant behaviors in malignancies in general and specifically in CRC.
References
Alexander, R. P., Fang, G., Rozowsky, J., Snyder, M. & Gerstein, M. B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 11, 559–571 (2010).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
Bernstein, B. E., Meissner, A. & Lander, E. S. The mammalian epigenome. Cell 128, 669–681 (2007).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Saito, Y., Liang, G., Egger, G., Friedman, J. M., Chuang, J. C., Coetzee, G. A. et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443 (2006).
Lujambio, A., Ropero, S., Ballestar, E., Fraga, M. F., Cerrato, C., Setien, F. et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429 (2007).
Brueckner, B., Stresemann, C., Kuner, R., Mund, C., Musch, T., Meister, M. et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 67, 1419–1423 (2007).
He, L., He, X., Lim, L. P., de Stanchina, E., Xuan, Z., Liang, Y. et al. A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 (2007).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004).
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 99, 15524–15529 (2002).
Calin, G. A., Liu, C. G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C. D. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA 101, 11755–11760 (2004).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 (2006).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 (2006).
Croce, C. M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714 (2009).
Calin, G. A., Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S. E. et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353, 1793–1801 (2005).
Iliou, M. S., da Silva-Diz, V., Carmona, F. J., Ramalho-Carvalho, J., Heyn, H., Villanueva, A. et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene 33, 4003–4015 (2014).
Fabbri, M., Garzon, R., Cimmino, A., Liu, Z., Zanesi, N., Callegari, E. et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3 A and 3B. Proc. Natl Acad. Sci. USA 104, 15805–15810 (2007).
Garzon, R., Liu, S., Fabbri, M., Liu, Z., Heaphy, C. E., Callegari, E. et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 113, 6411–6418 (2009).
Baer, C., Claus, R., Frenzel, L. P., Zucknick, M., Park, Y. J., Gu, L. et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 72, 3775–3785 (2012).
Corney, D. C., Flesken-Nikitin, A., Godwin, A. K., Wang, W. & Nikitin, A. Y. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 67, 8433–8438 (2007).
Mraz, M., Malinova, K., Kotaskova, J., Pavlova, S., Tichy, B., Malcikova, J. et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 23, 1159–1163 (2009).
Zenz, T., Mohr, J., Eldering, E., Kater, A. P., Buhler, A., Kienle, D. et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 113, 3801–3808 (2009).
Siemens, H., Jackstadt, R., Hunten, S., Kaller, M., Menssen, A., Gotz, U. et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10, 4256–4271 (2011).
Hahn, S., Jackstadt, R., Siemens, H., Hunten, S. & Hermeking, H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 32, 3079–3095 (2013).
Siemens, H., Neumann, J., Jackstadt, R., Mansmann, U., Horst, D., Kirchner, T. et al. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 19, 710–720 (2013).
Rokavec, M., Oner, M. G., Li, H., Jackstadt, R., Jiang, L., Lodygin, D. et al. IL-6 R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 124, 1853–1867 (2014).
Lujambio, A., Calin, G. A., Villanueva, A., Ropero, S., Sanchez-Cespedes, M., Blanco, D. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA 105, 13556–13561 (2008).
Toyota, M., Suzuki, H., Sasaki, Y., Maruyama, R., Imai, K., Shinomura, Y. et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68, 4123–4132 (2008).
Hill, L., Browne, G. & Tulchinsky, E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer 132, 745–754 (2013).
Feng, X., Wang, Z., Fillmore, R. & Xi, Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 344, 166–173 (2014).
Davalos, V., Moutinho, C., Villanueva, A., Boque, R., Silva, P., Carneiro, F. et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074 (2012).
Vrba, L., Jensen, T. J., Garbe, J. C., Heimark, R. L., Cress, A. E., Dickinson, S. et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE 5, e8697 (2010).
Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006).
Hur, K., Toiyama, Y., Takahashi, M., Balaguer, F., Nagasaka, T., Koike, J. et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 62, 1315–1326 (2013).
Lu, Y. X., Yuan, L., Xue, X. L., Zhou, M., Liu, Y., Zhang, C. et al. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin. Cancer Res. 20, 2631–2642 (2014).
Xi, Y., Formentini, A., Chien, M., Weir, D. B., Russo, J. J., Ju, J. et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights 2, 113–121 (2006).
Jerome, T., Laurie, P., Louis, B. & Pierre, C. Enjoy the silence: the story of let-7 microRNA and cancer. Curr Genomics 8, 229–233 (2007).
Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 (2004).
Dahiya, N., Sherman-Baust, C. A., Wang, T. L., Davidson, B., Shih Ie, M., Zhang, Y. et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE 3, e2436 (2008).
Roush, S. & Slack, F. J. The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516 (2008).
Lu, L., Katsaros, D., de la Longrais, I. A., Sochirca, O. & Yu, H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 67, 10117–10122 (2007).
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A. et al. RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005).
Kjersem, J. B., Ikdahl, T., Guren, T., Skovlund, E., Sorbye, H., Hamfjord, J. et al. Let-7 miRNA-binding site polymorphism in the KRAS 3'UTR; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5-fluorouracil and oxaliplatin +/- cetuximab. BMC Cancer 12, 534 (2012).
Chetty, R. & Govender, D. Gene of the month: KRAS. J. Clin. Pathol. 66, 548–550 (2013).
Markowitz, S. D. & Bertagnolli, M. M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 361, 2449–2460 (2009).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Mekenkamp, L. J., Tol, J., Dijkstra, J. R., de Krijger, I., Vink-Borger, M. E., van Vliet, S. et al. Beyond KRAS mutation status: influence of KRAS copy number status and microRNAs on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer 12, 292 (2012).
Ragusa, M., Majorana, A., Statello, L., Maugeri, M., Salito, L., Barbagallo, D. et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol. Cancer Ther. 9, 3396–3409 (2010).
Smits, K. M., Paranjape, T., Nallur, S., Wouters, K. A., Weijenberg, M. P., Schouten, L. J. et al. A let-7 microRNA SNP in the KRAS 3'UTR is prognostic in early-stage colorectal cancer. Clin. Cancer Res. 17, 7723–7731 (2011).
Zhang, W., Winder, T., Ning, Y., Pohl, A., Yang, D., Kahn, M. et al. A let-7 microRNA-binding site polymorphism in 3'-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann. Oncol. 22, 104–109 (2011).
Langevin, S. M. & Christensen, B. C. Let-7 microRNA-binding-site polymorphism in the 3'UTR of KRAS and colorectal cancer outcome: a systematic review and meta-analysis. Cancer Med. 3, 1385–1395 (2014).
Han, H. B., Gu, J., Zuo, H. J., Chen, Z. G., Zhao, W., Li, M. et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 226, 544–555 (2012).
Svoboda, M., Sana, J., Fabian, P., Kocakova, I., Gombosova, J., Nekvindova, J. et al. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat. Oncol. 7, 195 (2012).
Ogata-Kawata, H., Izumiya, M., Kurioka, D., Honma, Y., Yamada, Y., Furuta, K. et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9, e92921 (2014).
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 (2006).
Tuddenham, L., Wheeler, G., Ntounia-Fousara, S., Waters, J., Hajihosseini, M. K., Clark, I. et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 580, 4214–4217 (2006).
Migliore, C., Martin, V., Leoni, V. P., Restivo, A., Atzori, L., Petrelli, A. et al. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin. Cancer Res. 18, 737–747 (2012).
Varambally, S., Cao, Q., Mani, R. S., Shankar, S., Wang, X., Ateeq, B. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008).
Friedman, J. M., Liang, G., Liu, C. C., Wolff, E. M., Tsai, Y. C., Ye, W. et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 69, 2623–2629 (2009).
Strillacci, A., Griffoni, C., Sansone, P., Paterini, P., Piazzi, G., Lazzarini, G. et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res. 315, 1439–1447 (2009).
Strillacci, A., Valerii, M. C., Sansone, P., Caggiano, C., Sgromo, A., Vittori, L. et al. Loss of miR-101 expression promotes Wnt/beta-catenin signalling pathway activation and malignancy in colon cancer cells. J. Pathol. 229, 379–389 (2013).
Chandramouli, A., Onyeagucha, B. C., Mercado-Pimentel, M. E., Stankova, L., Shahin, N. A., LaFleur, B. J. et al. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol. Ther. 13, 175–183 (2012).
Duursma, A. M., Kedde, M., Schrier, M., le Sage, C. & Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 14, 872–877 (2008).
Zhang, H., Li, Y., Huang, Q., Ren, X., Hu, H., Sheng, H. et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 18, 1702–1710 (2011).
Chen, Y., Song, Y., Wang, Z., Yue, Z., Xu, H., Xing, C. et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J. Gastrointest. Surg. 14, 1170–1179 (2010).
Washietl, S., Findeiss, S., Muller, S. A., Kalkhof, S., von Bergen, M., Hofacker, I. L. et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA 17, 578–594 (2011).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Navarro, P., Page, D. R., Avner, P. & Rougeulle, C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. 20, 2787–2792 (2006).
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Plath, K., Fang, J., Mlynarczyk-Evans, S. K., Cao, R., Worringer, K. A., Wang, H. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003).
Yildirim, E., Kirby, J. E., Brown, D. E., Mercier, F. E., Sadreyev, R. I., Scadden, D. T. et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742 (2013).
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009).
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009).
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010).
Leveille, N., Elkon, R., Davalos, V., Manoharan, V., Hollingworth, D., Oude Vrielink, J. et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2, 513 (2011).
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Wu, Y., Zhang, L., Wang, Y., Li, H., Ren, X., Wei, F. et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 35, 9531–9538 (2014).
Li, L., Liu, B., Wapinski, O. L., Tsai, M. C., Qu, K., Zhang, J. et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 5, 3–12 (2013).
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011).
Graham, L. D., Pedersen, S. K., Brown, G. S., Ho, T., Kassir, Z., Moynihan, A. T. et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer 2, 829–840 (2011).
Ji, P., Diederichs, S., Wang, W., Boing, S., Metzger, R., Schneider, P. M. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B. & Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007).
Garen, A. & Song, X. Regulatory roles of tumor-suppressor proteins and noncoding RNA in cancer and normal cell functions. Int. J. Cancer. 122, 1687–1689 (2008).
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J. D. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 (2011).
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Wilusz, J. E., Freier, S. M. & Spector, D. L. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932 (2008).
Xu, C., Yang, M., Tian, J., Wang, X. & Li, Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int. J. Oncol. 39, 169–175 (2011).
Zheng, H. T., Shi, D. B., Wang, Y. W., Li, X. X., Xu, Y., Tripathi, P. et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 7, 3174–3181 (2014).
Morris, K. V. & Vogt, P. K. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle 9, 2544–2547 (2010).
Yu, W., Gius, D., Onyango, P., Muldoon-Jacobs, K., Karp, J., Feinberg, A. P. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206 (2008).
Pasmant, E., Laurendeau, I., Heron, D., Vidaud, M., Vidaud, D. & Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969 (2007).
Yap, K. L., Li, S., Munoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 (2010).
Kotake, Y., Nakagawa, T., Kitagawa, K., Suzuki, S., Liu, N., Kitagawa, M. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30, 1956–1962 (2011).
Thurman, R. E., Rynes, E., Humbert, R., Vierstra, J., Maurano, M. T., Haugen, E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant number 15K10108).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Kita, Y., Yonemori, K., Osako, Y. et al. Noncoding RNA and colorectal cancer: its epigenetic role. J Hum Genet 62, 41–47 (2017). https://doi.org/10.1038/jhg.2016.66
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.66
This article is cited by
-
Insights into the regulatory role of RNA methylation modifications in glioma
Journal of Translational Medicine (2023)
-
Long noncoding RNA Meg3 sponges miR-708 to inhibit intestinal tumorigenesis via SOCS3-repressed cancer stem cells growth
Cell Death & Disease (2021)
-
Epigenetics of colorectal cancer: biomarker and therapeutic potential
Nature Reviews Gastroenterology & Hepatology (2020)
-
Directional association test reveals high-quality putative cancer driver biomarkers including noncoding RNAs
BMC Medical Genomics (2019)
-
Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development
Genome Medicine (2019)