Abstract
Amyotrophic lateral sclerosis (ALS) is an intractable disease that causes respiratory failure leading to mortality. The main locus of ALS is motor neurons. The success of antisense oligonucleotide (ASO) therapy in spinal muscular atrophy (SMA), a motor neuron disease, has triggered a paradigm shift in developing ALS therapies. The causative genes of ALS and disease-modifying genes, including those of sporadic ALS, have been identified one after another. Thus, the freedom of target choice for gene therapy has expanded by ASO strategy, leading to new avenues for therapeutic development. Tofersen for superoxide dismutase 1 (SOD1) was a pioneer in developing ASO for ALS. Improving protocols and devising early interventions for the disease are vital. In this review, we updated the knowledge of causative genes in ALS. We summarized the genetic mutations identified in familial ALS and their clinical features, focusing on SOD1, fused in sarcoma (FUS), and transacting response DNA-binding protein. The frequency of the C9ORF72 mutation is low in Japan, unlike in Europe and the United States, while SOD1 and FUS are more common, indicating that the target mutations for gene therapy vary by ethnicity. A genome-wide association study has revealed disease-modifying genes, which could be the novel target of gene therapy. The current status and prospects of gene therapy development were discussed, including ethical issues. Furthermore, we discussed the potential of axonal pathology as new therapeutic targets of ALS from the perspective of early intervention, including intra-axonal transcription factors, neuromuscular junction disconnection, dysregulated local translation, abnormal protein degradation, mitochondrial pathology, impaired axonal transport, aberrant cytoskeleton, and axon branching. We simultaneously discuss important pathological states of cell bodies: persistent stress granules, disrupted nucleocytoplasmic transport, and cryptic splicing. The development of gene therapy based on the elucidation of disease-modifying genes and early intervention in molecular pathology is expected to become an important therapeutic strategy in ALS.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease (MND) among adults [1, 2]. No treatment, other than symptomatic management for dysphagia and respiratory failure, has been established. The pathomechanism of ALS has been elucidated by functional analysis of genes identified in familial ALS, which occurs in approximately 10% of patients with ALS. As many disease susceptibility genes have been reported in recent studies, genetic factors are now considered significantly involved in sporadic ALS [3].
Spinal muscular atrophy (SMA) is a motor neuron disease caused by a decrease in the survival motor neuron (SMN) protein due to SMN1 deficiency. Although SMN2, which is almost identical to SMN1, is present in vivo, mRNA from the gene is usually skipped in exon 7 due to splicing, and little functional SMN is synthesized. Nusinersen is a chemically-modified RNA that targets intronic splicing silencer N1 in intron 7 of SMN2 and inhibits the skipping of exon 7, thereby allowing the synthesis of functional SMN proteins from SMN2 [4]. In a randomized, double-blind, placebo-controlled trial (ENDEAR trial: NCT02193074) involving patients with infantile-onset SMA (type 1), nusinersen-treated patients showed a significant reduction in mortality and improvement in motor function [5]. Moreover, nusinersen has been shown to be effective in treating type 2 and type 3 SMA (CHERISH trial: NCT02292537) [6].
The approval of antisense oligonucleotides (ASOs) for treating SMA has had a significant impact and has brought hope to the development of therapies for other MNDs. Tofersen for superoxide dismutase 1 (SOD1) was a pioneer in developing ASO therapies for ALS [7]; however, it failed to show significant improvement in the primary endpoint in a phase 3 trial [8]. Therefore, improving protocols and developing early interventions for the disease are vital.
In this review, we have updated the knowledge of causative genes in ALS. Moreover, we have summarized the genetic mutations identified in familial ALS and their clinical features, focusing on SOD1, fused in sarcoma (FUS), and transacting response DNA-binding protein (TARDBP). A genome-wide association study (GWAS) has revealed disease-modifying genes that could be the target of gene therapy. The current status and prospects of gene therapy development were discussed, including ethical issues. Furthermore, we have discussed the potential of axonal pathology as a new therapeutic target from the perspective of early intervention.
The elucidation of causative genes has advanced our understanding of the pathogenesis of ALS, and genetic analysis has become an essential tool for developing personalized treatment
More than 30 ALS-causing and related genes have been identified
The identification of the SOD1 gene as the causative gene in 1993 was a major step forward in the study field of ALS [9,10,11]. Moreover, the TARDBP and FUS genes have been identified, and RNA metabolism has become a focus of attention as a pathological factor in ALS [12,13,14,15]. In 2001, ALS2/Alsin was identified as the causative gene for a young-onset autosomal recessive form of ALS [16]. In 2010, optineurin was identified as a cause of ALS with slow progression, extended duration, lower limb onset with spasticity, and cognitive impairment [17]. Both alsin and optineurin are autophagy-related molecules, suggesting that disruption of the protein degradation machinery is involved in the pathogenesis of ALS [18, 19].
In 2011, chromosome 9 open reading frame 72 (C9ORF72), the most frequent causative gene in familial ALS in Europe and the United States, was identified, which markedly changed the research trend [20, 21]. A C9ORF72 mutation has been described as frontotemporal dementia-ALS 1 (FTDALS1), which is found in approximately 40% of patients with familial ALS and 3% of patients with sporadic ALS in Europe and the United States [22]. Targeted resequencing and exome analysis of familial ALS using next-generation sequencers have been reported one after another and revealed novel causative genes [23,24,25,26]. As of December 2021, the Online Mendelian Inheritance in Man (OMIM) has registered 26 types of ALS and 24 causative genes, except for ALS3 and ALS7 (Table 1). ALS13, ALS24, and ALS25 have been suggested to be susceptibility genes (# in Table 1).
Racial/ethnic differences in genetic analysis
How often are these genetic mutations found in familial and sporadic ALS in Japan? The Japanese Consortium of ALS Research (JaCALS) has found known mutations in 48.7% of 39 Japanese families with suspected familial ALS, mainly in the autosomal dominant form, and known ALS-causing gene mutations in 3% of 469 cases of sporadic ALS [27]. We have been studying familial ALS since 1991. We performed targeted resequencing analysis of 111 Japanese families with suspected autosomal dominant forms of ALS [28]. We identified SOD1 mutations in 36 families, FUS mutations in 12 families, TARDBP mutations in two families, and optineurin p.E478G mutations in one family. We identified known mutations in 50% of the families with familial ALS [28].
The results of the analysis of causative genes in European and Japanese are shown in Fig. 1. Mutations in SOD1, TARDBP, FUS, and C9ORF72 were color-coded according to a review of the literature [27, 29], and data in Japan is modified from our previous study [28]. Mutations not determined (ND) in these four genes were also color-coded. Mutations were identified in 55.5% of Europeans with familial ALS and 43.6% of Japanese individuals with familial ALS. In sporadic ALS, mutations were also identified in 7.4% of European cases and 2.9% of Japanese cases. The difference between European and Japanese cases is mostly due to the difference in the frequency of C9ORF72 mutation. Furthermore, SOD1 and FUS are more common in Japanese, while TARDBP is more common in European. Asia represents over 50% of the world’s population; however, this continent is underrepresented in clinical trials and studies [30]. We want to point out that the frequency of the C9ORF72 mutation is low in Japan, unlike in Europe and the United States, while SOD1 and FUS are more common, indicating that the target mutations for therapy vary by ethnicity.
Racial/ethnic difference of amyotrophic lateral sclerosis (ALS) causative genes [28, 29, 93]. The pie charts of ALS causative genes in Europeans and Japan are shown, color-coded with SOD1, TARDBP, FUS, C9ORF72, and not determined (ND) in these four genes. Mutations were identified in 55.5% of Europeans with familial ALS and 43.6% of Japanese individuals with familial ALS. In sporadic ALS, only 7.4% of mutations were identified in Europe and 2.9% in Japan. The difference between European and Japanese is largely due to the difference in the frequency of C9ORF72 mutation, SOD1, and FUS being more common in Japanese and TARDBP being more common in European
Clinicogenetic and molecular characteristics of ALS genetic variants
SOD1
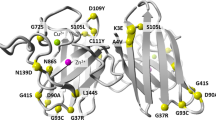
SOD1 (Cu/Zn-SOD) is a protein consisting of 153 amino acids, and more than 200 SOD1 mutations have been reported worldwide [9, 10]. Many cases are clinically indistinguishable from sporadic ALS, except for family history. The progression of disease with mutations of SOD1 is well correlated with each mutation [31, 32]. Most cases start in the lower motor neurons and lower limbs [33]. A correlation was found between certain point mutation and symptom severity, with p.A4V indicating severe disease and p.H46R indicating slow progression [34,35,36]. In our cohort, the mean age at onset was 48.4 years, and the mean disease duration was 4.9 years in patients with SOD1 mutations. The p.L126S mutation is characterized by rapid progression in homozygous cases [37] and relatively long course in heterozygous cases [38], with isolated inferior olivary hypertrophy in autopsy case [38]. The relatively frequent p.N86S mutation is characterized by phenotypic diversity and low penetrance even within families. p.L8V, a rare mutation that causes sensory disturbances, has also been reported [39]. Three mutations, p.H46R, p.L126S, and p.N86S, account for ~40% of the Japanese cases, with a lesser frequency of the severe form, p.A4V, which is estimated to account for 50% of SOD1 mutations among Europeans and Americans [40]. The p.D90A mutation is frequent in Europe [41], and is also transmitted in the AR form, but has not been found in our Japanese subjects [42]. These regional differences in SOD1 mutations should be taken into account in the development of SOD1-targeted therapies, such as the high prevalence of severe p.A4V in North America, p.D90A in Europe, and slower p.H46R in Japan [43, 44].
*The current HGVS nomenclature uses one amino acid shift compared to past nomenclature [45]. However, this review follows the traditional numbering for SOD1 variants in order to be consistent with previous articles.
FUS
In 2009, FUS was identified as the causative gene of ALS [12, 13]. FUS is the fourth most common causative gene for familial ALS in the US and Europe, following C9ORF72/SOD1/TARDBP. FUS mutation frequency is especially high in sporadic, early onset (<35 years of age) ALS patients because of de novo mutations [46]. In the original report, the average age at onset was 44.5 years and average survival was 33 months in familial ALS with FUS gene mutations, indicating early onset and rapid progress disease course [13].
In Japan, FUS mutations are the second most frequent ALS-causing gene mutations after SOD1 mutations in familial ALS [28, 47]. We have identified 11 mutations in 15 families, concentrated on Ex14, and 15 of FUS (Table 2) [48]. The FUS mutations tended to occur at a relatively young age, in the 30 or 40 s, with a cervical or upper extremity onset and a fast progressive disease course of ~2 years [48]. In contrast, families with the p.Q519E and p.S513P mutations had an older age of onset, a slightly slower progression rate, and a lower extremity onset. In a large family with the p.R521L mutation in the FUS gene, where we could obtain a detailed clinical profile of over five generations, 23 of 46 patients had ALS, and the penetrance rate was estimated to be as high as 100% [47]. At 35.3 years of age, the patients had muscle weakness, dysarthria, dysphagia, muscle spasticity, and atrophy [47]. The average age at death was 37.2 years, and the average time to need a ventilator was 23 months, indicating a young age of onset and a rapidly progressive course. In three autopsied cases with 1, 3, and 9 years of disease duration, the distribution of FUS-positive cytoplasmic inclusions according to disease stage was widespread [49]. In the case with a 9-year disease duration, in addition to the usual findings of ALS, histological examination revealed atrophy of the midbrain capsules, extensive neuronal loss of the substantia nigra, nucleus accumbens, subthalamic nucleus, and globus pallidus, especially the medial segment [49]. Cases with postural tremor and autism spectrum disorder were also reported [50].
TARDBP
In 2008, TARDBP (coding TDP-43) was identified as a causative gene of ALS [14, 15]. Before this finding, Arai et al. reported a pioneering biochemical and immunohistochemical analysis of TDP-43 inclusion bodies in autopsied brains of patients with ALS and FTD in 2006 [51]. In Japanese patients with ALS, TARDBP is the third most common familial ALS gene after SOD1 and FUS [28]. TDP-43 aggregation in the cytoplasm of spinal anterior horn neurons is a characteristic pathological finding in ALS and is observed in over 90% of patients with sporadic ALS and familial ALS with the C9ORF72 mutation. Considering the frequency of TARDBP mutations in familial ALS and the common TDP-43 pathology between sporadic ALS and familial ALS, focusing on TDP-43 in the pathogenesis of ALS is important. There are few reports of large families [15], and the penetration rate is considered low [52, 53].
Familial ALS with TARDBP mutations is more common in the limb onset and has a wider range of onset age [54]. Among these mutations, the p.G376D mutation had a particularly rapid progression, from onset to death in less than 1.5 years [52]. p.G298S mutation is also considered to be short-lived [55], whereas the p.A315T mutation had a longer disease course of 8–10 years. Focusing on rapidly progressing mutations is useful in pathological analysis using cellular and animal models, therefore clinical genetic analysis is important for understanding the pathogenesis of ALS.
C9ORF72
The frequency of the C9ORF72 mutation is high in Europe and the United States, indicating a founder effect of north European (Finnish) origin [56]. It forms a spectrum with FTD, as encoded by FTDALS1 (Table 1). The ALS-causing mutations in C9ORF72 are hexanucleotide repeat expansion in intron 1 [20, 21]. Bulbar onset has been more frequently observed in C9ORF72-mutated ALS [57]. It is still unclear whether anticipation also exists in C9orf72-associated diseases. Somatic and intergenerational repeat instabilities have been observed [57]. The disease penetrance of C9orf72-related ALS is thought to be nearly 100% by the age of 80 [58].
While the frequency of the C9ORF72 mutation is estimated to be 40% in Europe and the United States, it is less common in Japan, around 1–2% in familial ALS and 0.2% in sporadic ALS [59, 60]. The 20 SNPs in the North European consensus risk haplotype suggest a common ancestry. In the southern part of the Kii Peninsula, an area of high ALS prevalence in Japan, 3 of 15 cases were found to have C9ORF72 mutation [61]. In our own case, the patient had a typical ALS phenotype with onset between 50 and 70 years of age, frontotemporal dementia, and distal sensory deficits in the lower limbs.
Pathological hypotheses have been proposed: A. loss-of-function of C9ORF72; B. repeat-associated non-AUG (RAN) translation, which is a G4C2 repeat synthesized without the need for a transcription start site; and C. toxicity caused by a dipeptide repeat protein, which is synthesized by the translation of the G4C2 repeat [62]. The C9ORF72 protein is present in motor neurons [63]. C9ORF72 contributes to the maintenance of the immune environment, and its knockout is thought to cause abnormal immune responses, including the release of cytokines, associated with neurodegeneration [64]. Moreover, C9ORF72 knockout mice exhibited strikingly different survival rates depending on their environment and microbiome [65]. G4C2 repeats were transcribed as RNA and accumulated in the nucleus of nerve cells to form RNA foci by a liquid–liquid phase separation (LLPS) mechanism [66]. Furthermore, G4C2 repeat RNA and its translation product, dipeptide repeat protein, have been reported to cause neurodegeneration by increasing DNA double-strand breaks, leading to a deficiency of ataxia telangiectasia mutated (ATM), which repairs DNA damage [67]. Particularly, PR poly-dipeptides are highly toxic because they disrupt nucleocytoplasmic transport by polymerizing with the Nup54 protein in the nuclear pore [68]. A yeast study has found abnormalities in nucleocytoplasmic transport mediated by dipeptide repeat protein toxicity [69]. Furthermore, in a fly model overexpressing a hexanucleotide repeat, the phenotype caused by the C9ORF72 mutation was alleviated by increasing the expression of RAN GTPase-activating protein 1 (RanGAP1), a key regulator of nuclear and cytoplasmic transport [70]. The deletion of serine/arginine-rich splicing factor 1, which functions as an adapter during the transport of transcribed RNA out of the nucleus, has been reported to suppress neural degeneration by modulating RAN translation and could be a target for novel therapeutic agents [71]. Moreover, ASO, which inhibits repeat RNA, may be used as a therapeutic agent for ALS the C9ORF72 mutation, and GP repeat dipeptide in cerebrospinal fluid may be an alternative biomarker for determining drug efficacy [72].
Multisystem proteinopathy (MSP)
A group of patients known as inclusion body myopathy with Paget’s disease and FTD (IBMPFD) has been described [73, 74]. MSP, which is associated with FTD, inclusion body myopathy, and Paget’s disease of the bone in addition to ALS, has been recognized as an analogous disease concept [75, 76]. Recently, MSPs have been described in the OMIM as MSP1 with valosin-containing protein (VCP) mutation, MSP2 with the heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 mutation, MSP3 with the hnRNPA1 mutation, and MSP4 with the sequestosome-1 (SQSTM1)/p62 mutation [77]. The disease concept has been expanded to include matrin-3 mutations such as MSP5. Theoretically, any of the nearly 50 RNA-binding proteins with prion-like domains, such as FUS, could be a cause of MSPs [78]. hnRNPA1 mutations may cause only inclusion body myopathy in some families [79, 80]. Furthermore, genetic mutations associated with sensory impairment, parkinsonism, deafness, and various other allelic disorders are associated with MSPs (Table 1). MSP is an important disease concept in the context of motor neuron vulnerability and cell-specific pathology in ALS.
What GWAS has revealed
GWASs were conducted using single-nucleotide variant arrays to identify disease-associated variations in large cohorts of ALS cases and healthy controls. GWASs can reveal common genetic variants in thousands of unrelated individuals to identify associations with diseases that potentially explain certain percentages of disease heritability within a population [81]. The first GWAS in the ALS field was conducted in 2007, which highlighted the FLJ10986 gene as a candidate [82]. Following studies have revealed inositol 1,4,5-trisphosphate receptor type 2 [83], dipeptidyl peptidase like 6 [84], unc-13 homolog A (UNC13A) [85], Mps one binder kinase activator-like 2B [85], kinesin-associated protein 3 [86], cytochrome P450 family 27 subfamily A member 1 [87], zinc finger protein 512B [88], calcium/calmodulin-dependent protein kinase 1G [89], and sterile alpha and TIR motif-containing protein 1 [90] as representative ALS candidate genes. In 2015, the TANK-binding kinase 1 (TBK1) gene was identified as the causative gene for FTDALS4 in an analysis of 2,869 mainly sporadic ALS cases from groups centered in North America [91]. TBK1 gene variants are found in 1.26% of sporadic ALS in Japan including missense and loss-of-function mutations [92].
A GWAS using 1173 sporadic ALS cases and 8925 controls in a Japanese population combined with a meta-analysis of individuals of European ancestry has revealed a significant association at the Acyl-CoA synthetase long-chain family member 5 (ACSL5) locus [93]. A replication study involving a Chinese population and another set of the Japanese populations has confirmed the association. ACSL5 is involved in fatty acid metabolism, and other groups have found an association between ACSL5 single-nucleotide polymorphisms (SNPs) and lower fat-free mass in patients with ALS [94]. Serine palmitoyl transferase long-chain base subunit 1 is involved in the sphingolipid synthesis pathway and associated with juvenile ALS [95].
A recent cross-ancestry GWAS involving 29,612 patients with ALS and 122,656 controls identified 15 risk loci [96]. When combined with 6538 patients with whole-genome sequencing and a large cortex-derived expression quantitative trait locus dataset (MetaBrain), analyses have revealed locus-specific rare variants, short tandem repeats, and regulatory effects [96]. The combination of all ALS-associated signals reveals that perturbations contribute to vesicle-mediated transport and autophagy and provides evidence for cell-autonomous disease initiation in glutamatergic neurons [96]. Mendelian randomization analyses, which consider the environmental and lifestyle risk factors obtained from the literature, have indicated that high cholesterol levels play a causal role, again suggesting the importance of lipid metabolism [97]. In another recent analysis, machine learning RefMap method identified risk genes by integrating GWASs and epigenetic data [98]. Convergent genetic and experimental data revealed KN Motif And Ankyrin Repeat Domains 1 (KANK1) as a new ALS gene and initiation of ALS pathogenesis in the distal axon [98].
UNC13A is a gene repeatedly confirmed in several GWASs [99,100,101,102,103,104]. The C allele of the rs12608932 SNP within the UNC13A gene has been identified as a risk locus for both ALS and FTD [105]. Moreover, this SNP is associated with lower respiratory function at diagnosis and shorter survival [105]. Interestingly, a recent study has revealed that TDP-43 represses a cryptic exon-splicing event in UNC13A and reduces the expression of UNC13A [106, 107]. UNC13A contributes to vesicle priming and controls neurotransmitter release and short-term presynaptic plasticity [108]. UNC13A can be a stratification biomarker and a target of gene therapy. Although it should be noted that odds ratios are usually not high, many genes related to ALS have been identified using GWAS, and progress has been made in understanding the molecular pathogenesis of the disease (Table 3).
Oligogenic pathogenesis hypothesis
Up to this point, we have assumed a single gene mutation for a single patient. However, although SOD1 mutations are usually inherited in an autosomal dominant form with high penetrance, there are asymptomatic carriers of some types of mutations [28]. These observation has led to the idea that multiple causative genes cause the disease stage (oligogenic pathogenesis hypothesis), in which gene variants other than SOD1 are necessary for the onset of the disease [25]. More comprehensive panels of genetic testing will increase the possibility of detecting more than one rare variant in patients with ALS.
The identification of the C9ORF72 mutation in 2011 furthered this idea of oligogenic pathogenesis. For example, hexanucleotide repeat elongation of C9ORF72 with variants in other ALS causative genes are associated with a younger age of onset, suggesting that both mutations affect the onset of the disease [109]. Moreover, it can be viewed as a disease susceptibility gene, in the sense that both genetic variants affect the pathogenesis of the disease [28]. Whole-genome sequence of 4,315 cases revealed ALS-associated structural variants including inversion in the VCP gene and insertion in the ERBB4 gene [110]. Over 70% of respiratory onset ALS have ERBB4 insertion compared with 25% of the control [110]. Answer ALS project revealed 601 expanded regions in the 830 whole-genome sequence data using Expansion Hunter [111]. Large scale whole-genome open resources are now available.
There are other examples where multiple causative genes are associated with faster onset and progression of the disease [112,113,114]. Mutations in ataxin-2 cause a polyglutamine chain elongation of 34 repeats or more, which is a phenotype of spinocerebellar ataxia type 2. Ataxin-2 localizes to stress granules, and moderate repeat elongation promotes the activation of caspase 3, which produces TDP-43 C-terminus fragments, leading to ALS [115, 116]. Furthermore, there are ethnic differences in the intermediate-length CAG repeats of ataxin-2, with the “large normal allele” being less common in Japanese and more common in non-Japanese populations [117]. Ataxin-2 poly-CAG expansion is considered the target of ASO [118].
De novo mutations in sporadic ALS
The reported incidence of some ALS-associated variants in familial and sporadic ALS is different among causative genes. Moreover, 3–16% of sporadic ALS cases have monogenetic etiology (Fig. 1) [28, 29, 93, 119]. Others have reported that 21% of patients with ALS carried a confirmed pathogenic or likely pathogenic mutation, of whom 93% had no family history of ALS [120]. There could be several other cases of sporadic ALS with SOD1 mutations; however, either (a) DNA analysis of the parents showed one of them to be an asymptomatic mutation carrier; (b) the parents were not the biological parents, or (c) DNA was unavailable from one or both parents. A systematic review and meta-analysis has revealed that the estimated number of patients with SOD1 or C9ORF72 mutations are almost the same in familial ALS and sporadic ALS [121], suggesting that familial ALS classification based on reported family history does not capture the full picture of ALS of genetic origin [121].
Family history can be ambiguous or absent because of the following reasons: inadequate family history information in medical charts, misdiagnosis of ALS in older generations, reluctance to report hereditary disease, loss of contact between family members, low penetrance, small family size, early death due to other causes, development of ALS in offspring before the parent who transmitted the defective gene exhibits symptoms themselves, genetic pleiotropy, and lack of information on biological parents (i.e., adoption and illegitimacy) [122]. Moreover, 11.9% of patients carry a clinically relevant genetic mutation, and almost half of the reported mutations in the cohort has a prognostic value [123]. De novo mutations in FUS were reported in p.R495X and p.P525L cases [124, 125]; these mutations are also found in familial ALS cases. Moreover, de novo mutations in ataxin-2 [126], Erb-B2 receptor tyrosine kinase 4 [127], and Rap guanine nucleotide exchange factor 2 (RAPGEF2) have been reported in rare sporadic ALS cases [128]. Possible processes during embryogenesis of a de novo mutation in ALS could be the zygote, epiblast, and ectoderm. If the mutation happens in the ectoderm, we could not detect the mutation in the blood cells, confusing the interpretations.
The finding of de novo mutations in ALS provokes ethical implications. Patients with sporadic ALS who were genetically diagnosed vary in age (e.g., under 40 years old), in whether they have children, and by country (roughly one-third of the patients in Europe and the United States receiving genetic diagnosis) [129]. From the aspect of genetic counseling, it’s important to consider the facts that no fundamental treatment or prevention of the onset of the disease has been established, de novo mutations may be passed on to children or grandchildren, and that these facts might cause a large psychological burden. Providing sufficient explanation of the expected benefits and disadvantages before specimen collection and providing sufficient ethical consideration not only to the founder but also to their relatives are necessary. Moreover, collaborating with clinical geneticists and certified genetic counselors in explaining the results is important.
What is the state of the art in developing therapies targeting genetic mutations?
Advantages of ASO
The strategy of nusinersen was to compensate for the lack of SMN in SMA. Gene therapy includes introducing functional copies of dysfunctional genes, trophic factors, and disease-modifying genes or silencing the expression of harmful genes. Compared with other drug modalities (e.g., small-molecule or antibody drugs), nucleic acid medicine including ASO is a general term for drugs based on nucleic acids or artificial nucleic acids. Small-molecule drugs have the disadvantage that their drug targets are limited to receptors and enzymes. Antibody drugs are highly evaluated for their specificity and efficacy; however, their drug targets remain limited to molecules expressed on the cell membrane or secreted outside the cell, and mass production is difficult. Nucleic acid drugs have the advantages of being able to target intracellular target molecules, such as mRNA and non-coding RNA, which are difficult to target using conventional drugs, with high specificity and easy to manufacture.
The two main types of siRNAs have been invented: double-stranded RNAs, such as paticiran [130] approved for transthyretin amyloidosis, and single-stranded nucleic acids, such as ASOs. siRNAs currently under development for ALS are ASOs (Table 4). To target the central nervous system, ASOs are delivered either naked or by viral vectors, such as an adeno-associated virus (AAV) [131].
Current status of the therapeutic development of ASOs
Clinical trials of ASO are underway, focusing on SOD1 and C9ORF72, involving several patients. Gene therapy for SOD1 mutations has been attempted previously [132, 133]. Animal studies have been conducted to suppress the expression of mutant SOD1 by shRNA transduction using AAV vectors or by genome editing, both of which resulted in phenotypic improvement [132, 133]. Tofersen is an ASO drug being evaluated for the potential treatment of ALS with SOD1 mutations [7]. VALOR was a phase III, randomized, double-blind, placebo-controlled study that has evaluated the efficacy, safety, tolerability, and pharmacodynamic effects of tofersen on ALS with a confirmed SOD1 mutation (NCT02623699). Biogen and Ionis have reported that tofersen failed a first phase III trial to prove its effectiveness as the primary endpoint [8]. Early intervention with tofersen might be effective. Asymptomatic patients with known SOD1 mutations are eligible for the phase III ATLAS study, which examines the pre-symptomatic effect of tofersen, started in May 2021 (NCT0456982).
ASO treatment targeting C9ORF72, the most frequent target in Europe and the United States, has been investigated [134]. 245 AS101 is a phase I trial of BIIB078, which targets toxic RNA from hexanucleotide repeats while preserving normal C9ORF72 proteins, administered intrathecally to adults with C9ORF72-mutated ALS, sponsored by Biogen (NCT03626012). FOCUS-C9 is a phase Ib/IIa trial of the intrathecal administration of ASO (WVE-004-001) that promotes RNase H-mediated degradation of C9ORF72’s pathogenic mRNA variants associated with ALS or FTD and spares the normal C9ORF72 V2 variant in neurons (NCT04931862). Recently, the modification of a subset of a phosphodiester internucleotide linker is reported to improve ASO tolerability without impairing potency in repeated dosing for patients with C9ORF72 mutations [135], though additional clinical trials will be required to prove its efficacy.
The development of therapies for ASO that targets genes other than SOD1 and C9ORF72 is also progressing. ION363-CS1 is a phase I–III study that evaluates the efficacy and safety of intrathecally administering ION363/Jacifusen in ALS with FUS mutations. First-in-human treatment with ION363 silences FUS expression, decreases FUS pathology, and reverses insolubility of RNA-binding proteins in FUS-p.P525L mutated patients [136]. It was started in an n-of-1 trial but increased into an international 12-patient study at NEALS active sites (NCT04768972).
Since TDP-43 has been shown to be an important component of ubiquitin-positive and tau-negative inclusion bodies in most ALS cases, including sporadic ALS, the elucidation of the mechanism of abnormal TDP-43 aggregation has become a major research topic. TDP-43 is ubiquitously expressed and plays an important role in RNA metabolism and other cellular functions. Therefore, indirect methods for suppressing TDP-43 toxicity have been explored. For example, the knockdown of TDP-43 fails to maintain the number of motor neurons, and stathmin-2 (STMN2) becomes a mediator [137]. Selected ASOs have high tolerability in rodents but were not tested in monkeys; moderate potency in human motor neurons has been reported. An n-of-1 trial of STMN2 ASO was initiated. Initial doses were well tolerated. Although this is an ASO under research, it is the first human tolerability data on STMN2 (Symposium of ALSMND 2021 Dec).
The inhibition of ataxin-2 reduces the abnormal accumulation of TDP-43, prolongs survival, and improves motor function in mice overexpressing mutant TARDBP [118]. Moreover, ataxin-2 as an alternative target has attracted much attention [118]. 275AS101 is a phase I trial of BIIB105 targeting poly-CAG expansion in the ataxin-2 gene to reduce the ataxin-2 protein and mitigate TDP-43 toxicity [118] (NCT04494256).
ASO with constrained ethyl-group chemistry are partially absorbed from the gut [138]. Oral delivery can avoid lumbar-puncture-related adverse events like headaches. Conjugating cholesterol molecules to the ASO might also enhance penetration into the brain and spinal cord after systemic administration [139]. Oral ASO might be the feasible option.
Therapeutic development using AAV vectors
AAV-based therapies are under active development for various neuromuscular diseases [140]. AAV can be engineered for selective cell targeting and optimized transduction. Without the original viral genome, AAVs are nonpathogenic and unable to replicate like the wild-type virus. AAV9 penetrates the blood–brain barrier (BBB) and targets motor neurons to overcome an obstacle of gene therapy and reveals transduction with tropism with motor neurons [141]. In the case of SMA, onasemnogene abeparvovec (AVXS-101) could deliver across the BBB and into the spinal cord without integrating into the genome of the patient [142] and was approved worldwide. APB-102 is a recombinant AAVrh10 vector that expresses an anti-SOD1 artificial microRNA (Symposium of ALSMND 2021 Dec) (Table 4).
Other modalities include antibody drugs and small/medium molecule drugs
Drug discovery in modalities other than ASO is also becoming more active. Antibody drugs are being developed to target various causative gene products. AP-101 is a human monoclonal antibody targeting misfolded SOD1 generated by AL-S Pharma [143] and is now moving toward a human clinical trial. Misfolding-specific intrabody with dual proteolytic signals can eliminate TDP-43 inclusion [144]. Small molecules attached to ribonuclease reduced hexanucleotide repeat expansion in C9ORF72 mouse models [145]. Middle-sized peptides between small molecules and antibodies can be used for targeting “undruggable” intracellular protein–protein interactions [146, 147].
Studies have established the cellular phenotype of motor neurons using induced pluripotent stem cells (iPSCs) derived from patients with familial ALS [148, 149]. Screening of small molecules that improve the survival of motor neurons has also been reported, showing the efficacy of ropinirole and bosutinib, under the concept of drug repositioning [150,151,152]. A phase II trial of the potassium channel activator retigabine has indicated that short-interval intracortical inhibition as the primary outcome was significantly improved [153]. This work has shown how neurophysiological outcome measures could be used as disease markers.
Let us describe one more trial about developing drug for ALS. The establishment of the mutant SOD1 transgenic mouse model has greatly advanced research on the pathogenesis of ALS, but the approach to the brainstem and spinal cord, which are the main loci of the disease, has been limited by the small size of the mouse. To overcome the issue of sizing, we created a rat model of mutant SOD1 transgenes (ALS rats) [154]. The hepatocyte growth factor (HGF) is a novel growth factor originally cloned in Japan [155]. Overexpression of HGF in the nervous system attenuates disease progression and prolongs life span in a transgenic mouse model of ALS [156]. As for the idea of supplementing insufficient factors, intrathecal administration of human recombinant HGF (hrHGF) protein improved motor neuron pathology in a rat model with SOD1 mutation [154]. Phase II clinical trials are currently underway (UMIN000022050) [157].
The importance of rapid diagnosis and proper evaluation has been recognized in developing any therapeutic approach. Furthermore, the development of appropriate biomarkers and mechanisms to evaluate multiple therapeutic candidates is becoming increasingly important.
Innovations in biomarkers, clinical trial design, and definition of endpoints
Biomarkers are urgently needed for accurate stratification and diagnosis of patients with ALS for facilitating clinical trials. Moreover, neurofilament light (NFL) and phosphorylated neurofilament heavy chain (pNFH) are biomarkers for ALS [158, 159]. Plasma NFL levels are associated with a higher ALS risk in patients with pre-diagnostic ALS [160]. Plasma pNFH subunit levels are used as a secondary outcome of the trial of sodium phenylbutyrate and taurursodiol [161] or tofersen [7]. miR-181 was reported to have a prognostic value similar to that of NFL [162].
Using spinal cord samples from patients with sporadic ALS and ALS mouse models, vascular cell genes preceded the microglial response even at the pre-symptomatic stage [163]. Secreted phosphoprotein 1 (SPP1)- and COL6A1-positive perivascular fibroblasts accumulated in enlarged perivascular spaces in the spinal cord of patients with sporadic ALS. Increased levels of serum SPP1 could be a biomarker of shorter survival [163]. The combination of NFL and SPP1 or other markers, such as UNC13A genotype, might help stratify patients more effectively.
Combined endpoints have been used in several clinical trials to decrease the confounding effect of mortality on the analysis of functional outcomes, though survival and function are assessed as independent endpoints in ALS. The Combined Assessment of Function and Survival (CAFS) ranks patients’ clinical outcomes based on survival time and changes in the ALS Functional Rating Scale-Revised (ALSFRS-R) score [164]. Each patient’s outcome is compared with every other patient’s outcome, a score is assigned, and the summed scores are ranked. A higher mean CAFS score indicates a better group outcome. The CAFS endpoint was used as the primary endpoint of a dexpramipexole phase III study [165] and recent studies [166, 167].
Current clinical trial endpoints may not reflect what patients consider the most important and might estimate the benefit of novel treatments in the wrong way. A new composite endpoint for randomized controlled clinical trials of ALS named the Patient-Ranked Order of Function (PROOF), based on patient preference for functional domains is proposed [168].
In a recent systematic review, among 125 trials, investigating 76 drugs and recruiting more than 15,000 individuals with ALS, ~90% of trials have used traditional fixed designs [169]. To avoid resource limitations and barriers to trial participation in a rapidly progressive, disabling, and heterogeneous disease, a flexible and scalable multi-arm, the multi-stage trial platform is required.
The ethical concept of genetic testing must be discussed in view of drug development
We discussed de novo mutations in Section 1–6. In some reports, 21% of patients with ALS, of whom 93% had no family history, carried confirmed pathogenic, or likely pathogenic mutations [120]. Peripheral-blood exome, genome, and Sanger sequencing to identify pathologic mutations in SOD1 in 4000 patients with ALS from Germany, South Korea, and Sweden has revealed four sporadic ALS cases with de novo mutations in SOD1, which might be the therapeutic target of ASO [170]. To avoid missing the opportunity for treatment and earlier confirmed diagnosis, all patients with ALS should be offered genetic counseling and genetic screening. However, the challenges of variant interpretation associated with systematic genetic testing still exist; genetic testing must be accompanied by appropriate genetic counseling with human resources, variant interpretation, limited clinical trial spots, increased request for predictive testing, and psychosocial impact of identifying a genetic variant in patients without family history [129]. The International Consortium for Genetic Testing in ALS Committee was formed in March 2021, aiming to develop global best practice recommendations [129].
Understanding the molecular pathology for early therapeutic intervention: focus on axonal pathology (Fig. 2)
What are the pathological processes appropriate for early intervention? Axons are damaged early on
Overview of ALS pathology with a focus on axons. As axons are damaged from the initial stage of ALS, the dying-backward hypothesis, in which motor neurons are damaged from the distal part, has been proposed. RNA-seq of axon fraction shows the presence of intra-axonal transcription factors (e.g., AP-1), although the pathological significance remains unknown. NMJs are the key link between motor neurons and skeletal muscle, and NMJ disconnection is commonly observed in several types of ALS. The local translation is a molecular mechanism necessary for axonal homeostasis. In contrast, when the proteasome and autophagy are dysregulated, abnormal protein aggregation is triggered. Mitophagy is a form of autophagy, and mitochondrial pathology is a common feature of various neurodegenerative diseases. Furthermore, impaired axonal transport impairs the transport of RNA/Protein complex, lysosomes, and mitochondria. Many ALS-causing genes contribute to cytoskeleton function. Morphologically abnormal axon branching has been observed. The pathophysiology of the cell body, which is closely related to the axonal pathology, is also important. Cryptic exon induced nonsense-mediated decay (NMD) or aberrant proteins, persistent stress granules (SGs) formation, and nucleocytoplasmic transport defect have attracted attention as new therapeutic targets
As early/pre-symptomatic diagnosis becomes possible, as described in a previous chapter, intervening in the pathology becomes feasible at earlier stage. Looking back at the failure of the primary efficacy endpoint of a phase III trial of tofersen, interventions in the early stage are also indicated as critical [8]. In ALS animal models, morphological abnormalities of axons and neuromuscular junctions were observed from the early stages of the disease [171, 172]. Spastic paraplegia (SPG) type 11, profilin 1, never in mitosis gene A-related kinase 1 (NEK1), tubulin 4a, NFH, and chromosome 21 open reading frame 2 (C21ORF2 or CFAP410) have been identified as ALS-causing and susceptibility genes involved in axonal pathology and cytoskeletal abnormalities by ancestry analysis and GWASs [25, 100, 173]. C21ORF2 is mutated in ciliopathies [174] and is stabilized by NEK1-mediated hyperphosphorylation and the inability to bind F-box protein 3 [175]. Convergent genetic and experimental data revealed KANK1 as a new ALS gene and initiation of ALS pathogenesis in the distal axon [98].
Motor neurons are structurally characterized by long axons extending to the tips of the limbs. Focusing on the pathogenesis of both axon compartment and cell bodies may lead to the identification of new therapeutic targets. Mice with FUS mutation and hexanucleotide repeat expansion in C9ORF72 also showed axonal abnormalities, indicating a common pathology in ALS [171]. Abnormalities in axonal morphology and function, as well as the interaction with the extracellular environment to maintain the structure of long axons, are important pathologies that should be clarified as early and specific in motor neurons. In this chapter, we focused mainly on axonal pathology and considered its potential as a novel therapeutic target for ALS.
Axon sequencing reveals intra-axonal transcription factors
To understand what happens locally in axons, investigating the pathology of axon fraction itself is important. In case of cell culture setting, although several types of microfluidic devices are available on the market, some are specific to cell fraction analysis [176, 177], harvesting a sufficient sample for analysis from the axon compartment remained challenging [178]. A novel microfluidic device with improved dimensions of the well and materials enabled us to perform RNA sequencing using axon fraction (axon-seq) with fewer technical biases with the collection of several macroscopically observable axon bundles. Combining this innovative microfluidic device [179] with patient iPSC-derived motor neuron organoids further revealed the entire profile of the human motor neuron axons (Fig. 3) [180, 181]. This technique identified increased intra-axonal transcription factor, Fos-B (AP-1 family member) mRNA as a binding partner of FUS and as a causative event for aberrant axon morphology both in vitro and in vivo [181]. The upregulation of Fos-B mRNA is associated with increased spines [182, 183] and growth cones [184]. Activator protein-1 (AP-1) is increased in a mutant SOD1-G93A transgenic mouse model [185] and Fos-B protein accumulate abnormally in the motor neurons of sporadic ALS autopsy samples [181]; thus, dual leucine zipper kinase, the upstream signal protein for c-Jun (another AP-1 family member), and AP-1 family might become a common therapeutic target in ALS [185].
Motor neuron axons were extracted with a microfluidic device. a The experimental scheme of MN culture using the microfluidic device. HB9 reporter lentivirus infected motor precursor cells were plated onto the device. b Representative ICC images of MNs on the microfluidic device. The axons elongated in the microfluidics to the next well. c The enlarged images of nerve organoids stained with βIII-tubulin (cytoplasm) and Hoechst (nuclei). Bar: 1 mm. d Representative images of axon dividing. Axons were divided from the SDs by cutting the axon bundle at 450 μm away from the sphere to avoid contaminating the cell body and pushing out due to hydraulic pressure. Modified from ref. [181]
The data obtained may provide important resources for the subcellular fractional analysis of stem cell-derived motor neuron axons. The reproducibility of RNA profiles from the novel microfluidic device [176, 177] justified this approach. Notably, using TARDBP-mutated iPSCs, we found another intra-axonal transcription factor, paired-like homeobox protein 2B (phox2B), which showed a lower expression in mutant axons revealed by axon-seq and in situ hybridization [186]. Phox2B knockdown reduced neurite length in human and zebrafish motor neurons [186]. Phox2B-positive ocular motor neurons are resistant to degeneration in ALS compared with spinal motor neurons [187]. Targeted metabolomics identified elevated levels of the arachidonic acid pathway and reduction of arachidonic acid reverse ALS phenotypes in human and Drosophila spinal motor neurons and SOD1 mouse models [188].
Other groups have revealed the transcriptome of mature myelinated motor axons of the peripheral nervous system using the axon microdissection method devised by Koenig, enabling the isolation of axoplasm RNA to perform RNA-seq analysis [189]. The transcriptome analysis indicated the depletion of glial markers, enriched in neuronal markers and mRNAs related to the cytoskeleton, translation, and oxidative phosphorylation [189].
The whole story of what happens locally in the axon is unclear. Analyzing the molecular pathomechanism of axon fraction, including intra-axonal transcription factor, could provide a clue to elucidate the fragility of motor neurons in ALS.
Neuromuscular junctions (NMJs) are the key link between motor neurons and the effector skeletal muscle
ALS can be a distal axonopathy disease because many molecular changes of motor neuron degeneration start at NMJs [190]. The NMJ is a highly specialized synapse, which controls the signal between motor neurons and skeletal muscles. Clustering of acetylcholine receptors (AChR) at the NMJ are regulated by signaling molecules such as agrin, low-density lipoprotein receptor-related protein 4 (Lrp4), and muscle-specific receptor tyrosine kinase (MuSK) [191]. In a mutant SOD1-G37R transgenic mouse model, NMJ remodeling precedes the loss of the motor unit [192]. The activation of the muscle-specific kinase MuSK by the cytoplasmic protein Dok-7 is essential for NMJ formation, and Dok-7 recovery reduces muscle atrophy in a SOD1-G93A transgenic mouse model [193]. The upregulation of mitofusin 2 improves the NMJ morphology of mutant SOD1-G93A transgenic mice [194].
TDP-43 accumulation is observed in both intramuscular nerves of sporadic ALS and ALS patient iPSC-derived motor neuron axons [195]. Hyperphosphorylated TDP-43 promotes G3BP1-positive ribonucleoprotein condensate assembly and inhibits local protein synthesis in the axon and NMJ. Dissociation of G3BP1 condensates restores local translation and reduces toxicity by TDP-43 accumulation [195].
FUS mediates the transcriptional regulation of acetylcholine receptors at NMJs and is dysregulated in ALS [196]. Moreover, FUS is involved in NMJ maintenance and axonal transport [197, 198]. The expression of mutant FUS or FUS knockdown impairs motor activity and reduces acetylcholine transmission at NMJs in zebrafish [199]. Dipeptide repeat proteins related to C9ORF72 spread between motor neurons and skeletal muscles in vitro and in vivo [200]. Evidence suggests that the pathogenic mechanism of prion protein/exosome transfer is activated in the extracellular space and across the NMJ synapses during the degeneration of the motor cortex with centrifugal spreading [201,202,203].
Several NMJ microfluidic devices have been developed using human iPSC-derived motor neurons and myotubes [204]. An NMJ chip enables real-time, live imaging of axonal outgrowth, NMJ formation, and muscle maturation, as well as synchronization of motor neuron activity and muscle contraction under optogenetic control for analysis of physiological events [205]. Another important approach is the single-cell transcriptomics of nerve organoids in vitro [206]; pseudo-time analysis or single-cell trajectory analysis can help establish the relationship between the cause and effect of the transcriptome of the organoids [207, 208]. Sophisticated co-culture NMJ organoids can be beneficial for these studies.
Maintenance of axonal function by local translation
Whether mRNAs found in axon fractions are translated into axons or transported to the nucleus/cell body is an important question. Accumulating evidence has revealed that long motor neuron axons use asymmetrical mRNA localization and rely most strongly on mRNA transport and local translation to maintain homeostasis [209]. The anterior branch of human obturator motor neurons biopsied from patients with ALS demonstrated upregulation of a cluster of genes that play an important role in biological processes involving RNA processing and protein metabolism [210].
The upregulation of ribosome synthesis in axons occurs early in the pathogenesis of both mutant SOD1-G93A transgenic mouse models and human ALS autopsy samples, indicating aberrant axonal RNA metabolism [211]. Ribosomal protein mRNAs are locally translated and incorporated into native ribosomes in axons to maintain functional axonal ribosomes, which are reduced in sporadic ALS with TDP-43 pathology [212].
In SMA, reduced SMN decreases the axonal localization of several mRNAs [213] and inhibits the activity of the mammalian target of rapamycin in axons [214]. mTOR translationally alters the cytoskeletal regulator palladin to modulate axon morphogenesis [215]. Moreover, SMN regulates axonal localization and local translation of growth-associated protein 43 mRNA in growth cones through HuD and insulin-like growth factor 2 [216]. The aberrant distribution of SMN in cytosolic FUS accumulations reduces SMN in axons [217, 218]. The accumulation of mutant human FUS induces an integrated stress response and reduces protein synthesis in nearby axons [219]. Moreover, FUS mutation affects nonnuclear pools of splicing factor proline and glutamine-rich [220], which has been found to orchestrate spatial gene expression and is essential for axonal viability [221, 222].
Another example is casein kinase 2 alpha (CK2a). CK2a phosphorylates and triggers G3BP1 stress granule-like structure disassembly in injured axons [223]. CK2a activity is temporally and spatially regulated by the local translation of mRNA in axons after injury [223]. Axoplasmic calcium concentration is a determinant of the translational activation of different axonal mRNA and regenerative axonal growth [224].
Thus, local translation plays an important role in axonal homeostasis and nerve regeneration, which are dysfunctional in ALS.
Protein degradation: regulation of abnormal protein aggregation by proteasome and autophagy
The homeostatic processes engaged in eliminating defective organelles and aggregated proteins include autophagy and ubiquitin-proteasome systems. The accumulation of SOD1 and TDP-43 has been observed in autopsy specimens. Disruption of protein homeostasis has long been considered a pathogenic mechanism in ALS.
In the experimental models, constitutive autophagy in neurons maintains cellular homeostasis by balancing protein synthesis and degradation, particularly within the distal axonal processes [225, 226]. Disruption of the endosomal-lysosomal system by loss of Alsin deteriorates the phenotype of SOD1-H46R transgenic mice by accelerating the accumulation of misfolded proteins and immature vesicles in the spinal cord [19]. FUS mutation causes axonal retention of the FUS protein before its aggregation triggered by poly(ADP-ribose) polymerase-dependent DNA dependent repair signaling [227].
Optineurin is involved in autophagy and protein degradation pathways [17]. Optineurin binds to ubiquitin and regulates necrosis factor-kappa B activation and apoptosis [228]. Receptor-interacting kinase 1 (RIPK1)-dependent signaling is suppressed by optineurin by regulating its turnover [18]. Optineurin loss leads to progressive demyelination and axonal degeneration by activating necroptotic machinery in the central nervous system [18].
Moreover, optineurin is involved in several selective autophagy processes regulated by TBK1 [229]. TBK1 mutations are associated with impaired binding of autophagy adapter proteins. TBK1 phosphorylates and activates the Smith–Magenis syndrome chromosome region, candidate 8 (SMCR8), a member of the C9ORF72 complex, activating the autophagy pathway via RabGTPase [230]. FUS protein accumulation in autopsy cases with optineurin mutations [231] and decreased expression of TBK1 [232] suggest crosstalk between the disruption of protein homeostasis and abnormal RNA metabolism.
In GWAS, a meta-analysis of multiracial sporadic ALS data identified the GPX3-TNIP1 region, which encodes antioxidant glutathione peroxidase 3, and tumor necrosis factor-induced protein 3 interacting protein 1, a protein that interacts with optineurin [233]. Furthermore, mutations in the LC domain region of T cell-restricted intracellular antigen 1 suppress stress granule degradation and abnormal accumulation of TDP-43, which may be a cause of ALS [234]. Cyclin-F is a cell cycle regulator and ubiquitin E3 ligase [235] and interacts with SQSTM1/p62 in the autophagy pathway [236]. In zebrafish, a variant in UBQLN4 compromises motor axon morphogenesis, impairing proteasomal function [237, 238].
The identification of these genes indicates that the disruption of protein homeostasis is an important common mechanism in ALS, especially in the compartment of motor neuron axons.
Mitochondrial pathology
The mitochondria generate adenosine triphosphate through oxidative phosphorylation and provide the axonal energy demand [239]. After synthesis at the cell body, the mitochondria accumulate at the nodes of Ranvier to meet metabolic needs [240]. Several neurodegenerative diseases are affected by disrupted mitochondrial activity, transport proteins, and microtubule association [241]. Mutations in RAPGEF2 impair microtubule stability and mitochondrial distribution in axons [242]. Dysfunction in Rho GTPase 1 (Miro 1), the outer mitochondrial membrane protein, leads to anterograde axonal transport defects [243]. The imbalance between mitochondrial fission and fusion leads to abnormal morphology of the mitochondria in spastic paraparesis [244]. Syntaphilin, a mitochondria-anchoring protein, mediates the clearance of dysfunctional mitochondria from motor neuron axons [245]. Loss-of-function mutations in SIGMAR1 decrease mitochondria-associated membrane (MAM), impairing retrograde transport and axonal degeneration [246, 247].
Phosphatase and tensin homologs deleted from chromosome 10-induced kinase 1 (PINK1) and parkin are key regulators of mitophagy, a selective autophagic pathway to eliminate dysfunctional mitochondria [248]. The disruption of PINK1 signaling is found in SOD1 mutant mice and samples from patients with sporadic ALS [249]. Parkin expression is regulated by TDP-43 and reduced in motor neurons of TDP-43 pathology with ALS [250, 251]. Axonal transport of mitochondria is disrupted in FUS-mutant patients with aggregation of Parkin and PINK1 [252].
RNA-seq revealed reduced gene expression of mitochondrially encoded electron transport chain transcripts, and neuropathological analysis of C9ORF72-mutated ALS postmortem tissue confirmed selective dysregulation of mitochondrially encoded transcripts in ventral horn spinal motor neurons [253]. Genetic manipulation of mitochondrial biogenesis in C9ORF72 motor neurons corrected the bioenergetic deficit and rescued the axonal length and transport phenotypes [253].
The degradation of autophagic vacuoles that engulf damaged mitochondria is impaired in distal axons in a SOD1-G93A transgenic mouse model [254]. A potential drug that reduced neuronal cell death in a SOD1-G93A mouse model is a neuronal SIGMA1-receptor agonist (SA4503), which reduces oxidative stress and regulates calcium flux in the mitochondria [255]. Pridopidine, a SIGMA1 agonist, is being investigated in the HEALEY ALS Platform Trial (NCT04615923). Clinical trials of the combination of dextromethorphan and quinidine, which affect the demethylation of the P450 cytochrome enzyme, have revealed improvement in the pseudobulbar effect in patients with ALS [256]. Mitochondrial activation drugs, such as MA-5 [257], have potential as a strategy for various mitochondrial dysfunction pathologies.
Mitochondrial metabolism is the emerging and noteworthy therapeutic target in ALS.
Axonal transport
Intracellular transport of cargo is especially important in neurons because of the polarization between axon and cell bodies [258]. RNA/protein complexes and organelles, such as the mitochondria, are synthesized in the soma and transported along the axon. The distribution of this cargo at the right time and place in the axon depends on the proper transportation of the cargo. The transport defect was revealed in ALS, and the proximal axons of large motor neurons harbor abnormal accumulation of mitochondria, phosphorylated neurofilaments, and lysosomes [259,260,261,262]. Furthermore, the structure of spheroids in motor nerve axons in ALS autopsy samples contains different types of vesicles, neurofilaments, lysosomes, mitochondria, and microtubules [258, 263], suggesting axonal transport reduction.
Aberrant cargo transport within axons occurs early in ALS disease progression in mutant SOD1-G93A transgenic mouse models of ALS [264,265,266]. Inhibiting p38 MAPK rescues retrograde cargo transport defects within axons of mutant SOD1-G93A transgenic mouse models [267]. Moreover, ALS-related mutations in TDP-43 alter the transport function [268]. Similarly, defects in cargo transport within axons were demonstrated in FUS-mutant iPSC-derived motor neurons, which were rescued by inhibiting histone deacetylase 6 [269].
Dynactin 1, which binds to microtubules, is a motor protein responsible for the retrograde transport of various proteins and vesicles [270]. ALS and slowly progressing, autosomal dominant, distal hereditary motor neuropathy in vocal paresis are due to loss-of-function mutations in dynactin 1 [271,272,273]. Kinesin is another motor protein involved in the anterograde axonal transport [274]. Its family member 5A (KIF5A) is mutated in the N-terminal motor domain in SPG10 and CMT type 2, whereas the C-terminal domain is mutated in ALS [275]. Patients with loss-of-function KIF5A mutations have longer survival times than those with typical ALS [276, 277]. Furthermore, loss-of-function mutations in KIF1A are present in the motor or neck domains [278, 279]. These motor proteins are dysregulated in sporadic ALS pathology [280]. Loss-of-function mutation in KIF5A is found in 0.12% of sporadic ALS in Japan [281].
Annexin A11, a phosphoinositide-binding protein associated with RNA granules, functions as a molecular tether between lysosomes and RNA granules. Such tethering is impaired by ALS-associated annexin A11 mutations [282, 283]. Late endosome-bearing mRNAs encoding mitochondrial functional molecules stop at the mitochondria, and these mRNAs are translated on Rab7a endosomes locally in the axon [284]. Axonal transport and other pathological processes, such as autophagy and mitophagy, closely interact. Defects in the cargo transport within axons are common in various neurodegenerative diseases [285].
Aberrant cytoskeleton and axon branching
Several variants of the gene encoding α-tubulin destabilize the microtubule network and reduce the repolymerization capability of the cytoskeleton [173]. Mutations in profilin 1, which converts monomeric actin to filamentous actin, lead to familial ALS. Ubiquitinated aggregates, including TDP-43, are present in cells that express mutant profilin 1 [286]. Reduced binding with actin and axon growth are observed in mutant profilin 1-expressing cells. Profilin 1 transgenic mice have been observed to recapitulate the phenotype of MNDs [287]. C9ORF72 modulates the activity of small GTPases, increases the activity of LIM kinases 1 and 2, and regulates axonal actin dynamics [288]. Various actin isoforms are expressed in primary mouse motor neurons, and their transcripts are translocated into motor neuron axons [289]. NFL transcripts are reduced in ALS [290]. Moreover, neurofilaments are found in a spheroid structure (large axonal swelling) [291]. NFL and pNFH are also known as biomarkers for ALS [158, 159]. Thus, cytoskeleton abnormal morphologies contribute to the pathogenesis of ALS. Non-labeled live imaging of stimulated Raman scattering microscopy [292] can visualize peripheral degeneration in live ALS mouse models and human postmortem tissue [171]. The novel technology might be a supportive tool for diagnosing cytoskeletal abnormalities much earlier and assessing the effectiveness of the therapies.
TDP-43 is a crucial splicing repressor, and its loss results in novel cryptic exons being erroneously included in mature mRNA. One of the splicing targets of TDP-43, STMN2, a regulator of microtubule stability, is involved in the pathomechanism of TARDBP mutations [137, 293]. STMN2 is decreased following TARDBP knockdown due to altered splicing when TDP-43 is mislocalized and in motor neurons from patients and the spinal cord of postmortem samples. Posttranslational STMN2 stabilization rescues neurite outgrowth and axon regeneration deficits by depleting TDP-43 [137]. QurAlis QRL-201 is a therapeutic ASO that restores STMN2 mis-splicing due to TDP-43 pathology.
Axonal branching is a key mechanism of synaptic plasticity [294]. Aberrant axonal branching is implicated in the pathomechanisms of ALS. For example, motor neurons cultured from mutant SOD1-G93A transgenic mouse models exhibit enhanced axonal branching [295]. The overexpression of mutant human TARDBP in zebrafish embryos induces a phenotype that includes shorter motor neuron axons, premature and increased axonal branching, and ends in abnormal swimming [296]. Progranulin rescues mutant TARDBP-induced aberrant axonal branching and short axonal outgrowth [297]. Branching in FUS-mutant motor neuron axons is increased compared with that in isogenic controls in vitro [181]. This phenotype was confirmed using other ALS causative mutations, including SOD1 and TARDBP [181]. Morphological changes in motor neuron axon branching have been found to precede motor neuron death in mutant SOD1-G93A transgenic mouse models [171], and abnormal neural branching has been detected in zebrafish that overexpress mutant FUS [199]. Moreover, other groups have reported increased axon branching in FUS, SOD1, and TARDBP mutant iPSC models [298]. SMN knockdown in zebrafish embryos significantly increases motor neuron branching [299]. Furthermore, mutant CCNF zebrafish developed a motor neuron axonopathy, which consists of shortened primary motor neuron axons and an increased frequency of aberrant axonal branching [235, 300].
The meaning of axonal branching might be different in each developmental stage [301]. In the embryonic stage, axon pathfinding and synaptic formation are important. However, in the developed stage, aberrant axon branching might have a disadvantage in terms of the normal function of electronic neurotransmission. The significance of aberrant axonal branching in the context of the neurodegenerative model in vivo must be elucidated.
The pathology of the cell body is also important: persistent stress granule formation and nucleocytoplasmic transport
Although we have focused on axonal pathology, considering the pathology occurring in motor neuron cell bodies is also important to understand the overall picture of ALS. FUS and TDP-43, which normally reside in the nucleus, have RNA-binding domains that regulate RNA metabolism and transport [302]. Mutant TDP-43 and FUS change their localization from the nucleus to the cytoplasm [303], suggesting that they gain or lose function in the nucleus [304] and are important in normal physiological aging [304]. Stress granules are droplets that form under various stresses, such as heat shock and hypoxia, and are composed of mRNA and RNA-binding proteins [305]. They protect cellular homeostasis by temporarily inactivating mRNA translation under stress and allowing it to resume after the stress is removed. TDP-43, FUS, hnRNPA1, and hnRNPA2 are nuclear RNA-binding proteins with prion-like domains and undergo LLPS to form functional liquids, including stress granules, which can be converted into abnormal hydrogels that contain pathological fibrils often seen in neurodegenerative diseases [306,307,308,309]. Mutations in prion-like domains increase the rate of fibril formation and initiate disease [310]. Karyopherin-β2 (also known as transportin-1) binds the proline-tyrosine nuclear localizing signal and then blocks and reverses FUS fibril formation [310,311,312,313]. Moreover, importin-α and karyopherin-β1 block and reverse TDP-43 fibril formation [310]. The identification of molecules involved in the assembly/disassembly of LLPS is being actively pursued because the identification of molecules that regulate the dissociation of LLPS droplets in cells may help avoid aggregation toxicity [314,315,316].
Alterations in nucleoporins and the function of nuclear pore complexes have also been considered a therapeutic target in ALS [317,318,319]. RanGAP1 regulates the directionality of the transport [320]. If RAN gradients are impaired, cytoplasmic accumulation of nuclear proteins, as well as defected export of mature RNAs from the nucleus, sequesters nucleocytoplasmic transport [319]. Endosomal sorting complexes required for transport machinery, including charged multivesicular body protein 7 (CHMP7), is involved in proteasomal degradation of disassembled nucleoporins [321]. Inhibiting the nuclear export of CHMP7 triggers nucleoporin reduction, and TDP-43 dysfunction and knockdown of CHMP7 alleviate Ran GTPase mis-localization [322]. Moreover, mutated FUS interacts with nucleoporins and declines nucleocytoplasmic transport in Drosophila and iPSCs [323]. Toxic proline:arginine dipeptides from C9ORF72 bind to karyopherin-β2 and impede nucleocytoplasmic transport by interacting with nuclear import receptors [324].
Dying-forward and dying-backward hypotheses are both important [325, 326]. The association between axonal dysfunction and these cell body/nuclei events, including nucleocytoplasmic transport and stress granule/aggregation formation, should be elucidated in the context of axonal pathology.
Conclusion
With the development of edaravone as the second ALS drug after riluzole, the treatment of ALS has made steady progress based on the knowledge gained from the elucidation of the pathogenesis of familial ALS. Comprehensive analyses of causative genes, disease susceptibility genes, and disease-modifying genes will continue to be important for the complete understanding of the disease. Targeting axons, the early pathological site of ALS, is also desirable. In the future, there is a strong need for developing more effective therapies to prevent neurodegeneration and symptomatic treatments to alleviate the disease. Like nusinersen for SMA, expectations for ASO remain high.
With the paradigm shift in therapeutic development, we must debate ethical issues, such as genetic diagnosis, in sporadic ALS. Because of the impact on the patient and the family, genetic diagnosis also places a heavy psychological burden on the attending physician. Even if a treatment is developed in the future, prenatal diagnosis will make it even more difficult to make ethical choices. Even if the guidelines can provide examples, each particular case will be subject to difficult decisions. It will also be necessary to take into accounts the mental exhaustion of not only the patient and family but also the medical profession.
References
Al-Chalabi A, van den Berg LH, Veldink J. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat Rev Neurol. 2017;13:96–104.
Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Prim. 2017;3:17085.
Gibson SB, Downie JM, Tsetsou S, Feusier JE, Figueroa KP, Bromberg MB, et al. The evolving genetic risk for sporadic ALS. Neurology. 2017;89:226–33.
Singh NN, Howell MD, Androphy EJ, Singh RN. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017;24:520–6.
Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, et al. Nusinersen versus Sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377:1723–32.
Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, et al. Nusinersen versus Sham control in later-onset spinal muscular atrophy. N Engl J Med. 2018;378:625–35.
Miller T, Cudkowicz M, Shaw PJ, Andersen PM, Atassi N, Bucelli RC, et al. Phase 1-2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2020;383:109–19.
Mullard A. ALS antisense drug falters in phase III. Nat Rev Drug Disco. 2021;20:883–5.
Aoki M, Ogasawara M, Matsubara Y, Narisawa K, Nakamura S, Itoyama Y, et al. Mild ALS in Japan associated with novel SOD mutation. Nat Genet. 1993;5:323–4.
Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62.
Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–51.
Kwiatkowski TJ Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8.
Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11.
Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–4.
Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72.
Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–73.
Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6.
Ito Y, Ofengeim D, Najafov A, Das S, Saberi S, Li Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–8.
Hadano S, Otomo A, Kunita R, Suzuki-Utsunomiya K, Akatsuka A, Koike M, et al. Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PLos ONE. 2010;5:e9805.
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56.
Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68.
Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23.
Dekker AM, Seelen M, van Doormaal PT, van Rheenen W, Bothof RJ, van Riessen T, et al. Large-scale screening in sporadic amyotrophic lateral sclerosis identifies genetic modifiers in C9orf72 repeat carriers. Neurobiol Aging. 2016;39:e9–15.
Chia R, Chio A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17:94–102.
Nguyen HP, Van Broeckhoven C, van der Zee J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018;34:404–23.
Mathis S, Goizet C, Soulages A, Vallat JM, Le, Masson G. Genetics of amyotrophic lateral sclerosis: a review. J Neurological Sci. 2019;399:217–26.
Nakamura R, Sone J, Atsuta N, Tohnai G, Watanabe H, Yokoi D, et al. Next-generation sequencing of 28 ALS-related genes in a Japanese ALS cohort. Neurobiol Aging. 2016;39:e1–8.
Nishiyama A, Niihori T, Warita H, Izumi R, Akiyama T, Kato M, et al. Comprehensive targeted next-generation sequencing in Japanese familial amyotrophic lateral sclerosis. Neurobiol Aging. 2017;53:e1–8.
Zou ZY, Zhou ZR, Che CH, Liu CY, He RL, Huang HP. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2017;88:540–9.
Shahrizaila N, Sobue G, Kuwabara S, Kim SH, Birks C, Fan DS, et al. Amyotrophic lateral sclerosis and motor neuron syndromes in Asia. J Neurol Neurosur Psychiatry. 2016;87:821–30.
Aoki M, Abe K, Itoyama Y. Molecular analyses of the Cu/Zn superoxide dismutase gene in patients with familial amyotrophic lateral sclerosis (ALS) in Japan. Cell Mol Neurobiol. 1998;18:639–47.
Aoki M, Abe K, Houi K, Ogasawara M, Matsubara Y, Kobayashi T, et al. Variance of age at onset in a Japanese family with amyotrophic lateral sclerosis associated with a novel Cu/Zn superoxide dismutase mutation. Ann Neurol. 1995;37:676–9.
Bernard E, Pegat A, Svahn J, Bouhour F, Leblanc P, Millecamps S, et al. Clinical and molecular landscape of ALS patients with SOD1 mutations: novel pathogenic variants and novel phenotypes. A single ALS Center Study. Int J Mol Sci. 2020;21:6807.
Saeed M, Yang Y, Deng HX, Hung WY, Siddique N, Dellefave L, et al. Age and founder effect of SOD1 A4V mutation causing ALS. Neurology. 2009;72:1634–9.
Zou ZY, Liu MS, Li XG, Cui LY. H46R SOD1 mutation is consistently associated with a relatively benign form of amyotrophic lateral sclerosis with slow progression. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17:610–3.
Aoki M, Ogasawara M, Matsubara Y, Narisawa K, Nakamura S, Itoyama Y, et al. Familial amyotrophic lateral sclerosis (ALS) in Japan associated with H46R mutation in Cu/Zn superoxide dismutase gene: a possible new subtype of familial ALS. J Neurol Sci. 1994;126:77–83.
Kato M, Aoki M, Ohta M, Nagai M, Ishizaki F, Nakamura S, et al. Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci Lett. 2001;312:165–8.
Hideshima M, Beck G, Yamadera M, Motoyama Y, Ikenaka K, Kakuda K, et al. A clinicopathological study of ALS with L126S mutation in the SOD1 gene presenting with isolated inferior olivary hypertrophy. Neuropathology. 2020;40:191–5.
Nishiyama A, Warita H, Takahashi T, Suzuki N, Nishiyama S, Tano O, et al. Prominent sensory involvement in a case of familial amyotrophic lateral sclerosis carrying the L8V SOD1 mutation. Clin Neurol Neurosurg. 2016;150:194–6.
Bali T, Self W, Liu J, Siddique T, Wang LH, Bird TD, et al. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017;88:99–105.
Berdynski M, Miszta P, Safranow K, Andersen PM, Morita M, Filipek S, et al. SOD1 mutations associated with amyotrophic lateral sclerosis analysis of variant severity. Sci Rep. 2022;12:103.
Pereira GRC, Vieira BAA, De Mesquita JF. Comprehensive in silico analysis and molecular dynamics of the superoxide dismutase 1 (SOD1) variants related to amyotrophic lateral sclerosis. PLos ONE. 2021;16:e0247841.
Radunovic A, Leigh PN. Cu/Zn superoxide dismutase gene mutations in amyotrophic lateral sclerosis: correlation between genotype and clinical features. J Neurol Neurosurg Psychiatry. 1996;61:565–72.
Cudkowicz ME, McKenna-Yasek D, Chen C, Hedley-Whyte ET, Brown RH Jr. Limited corticospinal tract involvement in amyotrophic lateral sclerosis subjects with the A4V mutation in the copper/zinc superoxide dismutase gene. Ann Neurol. 1998;43:703–10.
den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–9.
Hubers A, Just W, Rosenbohm A, Muller K, Marroquin N, Goebel I, et al. De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiol Aging. 2015;36:3117. e1–17.e6.
Suzuki N, Aoki M, Warita H, Kato M, Mizuno H, Shimakura N, et al. FALS with FUS mutation in Japan, with early onset, rapid progress and basophilic inclusion. J Hum Genet. 2015;55:252–4.
Akiyama T, Warita H, Kato M, Nishiyama A, Izumi R, Ikeda C, et al. Genotype-phenotype relationships in familial amyotrophic lateral sclerosis with FUS/TLS mutations in Japan. Muscle Nerve. 2016;54:398–404.
Suzuki N, Kato S, Kato M, Warita H, Mizuno H, Kato M, et al. FUS/TLS-immunoreactive neuronal and glial cell inclusions increase with disease duration in familial amyotrophic lateral sclerosis with an R521C FUS/TLS mutation. J Neuropathol Exp Neurol. 2015;71:779–88.
Eura N, Sugie K, Suzuki N, Kiriyama T, Izumi T, Shimakura N, et al. A juvenile sporadic amyotrophic lateral sclerosis case with P525L mutation in the FUS gene: a rare co-occurrence of autism spectrum disorder and tremor. J Neurol Sci. 2019;398:67–8.
Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11.
Mitsuzawa S, Akiyama T, Nishiyama A, Suzuki N, Kato M, Warita H, et al. TARDBP p.G376D mutation, found in rapid progressive familial ALS, induces mislocalization of TDP-43. eNeurologicalSci. 2018;11:20–2.
Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193.
Harms MB, Baloh RH. Clinical neurogenetics: amyotrophic lateral sclerosis. Neurol Clin. 2013;31:929–50.
Corcia P, Valdmanis P, Millecamps S, Lionnet C, Blasco H, Mouzat K, et al. Phenotype and genotype analysis in amyotrophic lateral sclerosis with TARDBP gene mutations. Neurology. 2012;78:1519–26.
Smith BN, Newhouse S, Shatunov A, Vance C, Topp S, Johnson L, et al. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. Eur J Hum Genet. 2013;21:102–8.
Hubers A, Marroquin N, Schmoll B, Vielhaber S, Just M, Mayer B, et al. Polymerase chain reaction and Southern blot-based analysis of the C9orf72 hexanucleotide repeat in different motor neuron diseases. Neurobiol Aging. 2014;35:1214.e1–6.
Volk AE, Weishaupt JH, Andersen PM, Ludolph AC, Kubisch C. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Med Genet. 2018;30:252–8.
Konno T, Shiga A, Tsujino A, Sugai A, Kato T, Kanai K, et al. Japanese amyotrophic lateral sclerosis patients with GGGGCC hexanucleotide repeat expansion in C9ORF72. J Neurol Neurosurg Psychiatry. 2013;84:398–401.
Ogaki K, Li Y, Atsuta N, Tomiyama H, Funayama M, Watanabe H, et al. Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:e11–6.
Ishiura H, Takahashi Y, Mitsui J, Yoshida S, Kihira T, Kokubo Y, et al. C9ORF72 repeat expansion in amyotrophic lateral sclerosis in the Kii peninsula of Japan. Arch Neurol. 2012;69:1154–8.
Taylor JP, Brown RH Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206.
Suzuki N, Maroof AM, Merkle FT, Koszka K, Intoh A, Armstrong I, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16:1725–7.
Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med. 2016;8:347ra93.
Burberry A, Wells MF, Limone F, Couto A, Smith KS, Keaney J, et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature. 2020;582:89–94.
Nedelsky NB, Taylor JP. Pathological phase transitions in ALS-FTD impair dynamic RNA-protein granules. RNA. 2022;28:97–113.
Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci. 2017;20:1225–35.
Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci USA. 2017;114:E1111–7.
Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–9.
Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61.
Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, et al. SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat Commun. 2017;8:16063.
Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, et al. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:eaai7866.
Kovach MJ, Waggoner B, Leal SM, Gelber D, Khardori R, Levenstien MA, et al. Clinical delineation and localization to chromosome 9p13.3-p12 of a unique dominant disorder in four families: hereditary inclusion body myopathy, Paget disease of bone, and frontotemporal dementia. Mol Genet Metab. 2001;74:458–75.
Kimonis V. Inclusion body myopathy with paget disease of bone and/or frontotemporal dementia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al., editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993–2022.
Taylor JP. Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology. 2015;85:658–60.
Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73.
Benatar M, Wuu J, Fernandez C, Weihl CC, Katzen H, Steele J, et al. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology. 2013;80:1874–80.
Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417–38.
Izumi R, Warita H, Niihori T, Takahashi T, Tateyama M, Suzuki N, et al. Isolated inclusion body myopathy caused by a multisystem proteinopathy-linked hnRNPA1 mutation. Neurol Genet. 2015;1:e23.
Suzuki N, Aoki M, Mizuno H, Onodera Y, Takahashi T, Nagata T, et al. Late-onset distal myopathy with rimmed vacuoles without mutation in the GNE or dysferlin genes. Muscle Nerve. 2005;32:812–4.
Rich KA, Roggenbuck J, Kolb SJ. Searching far and genome-wide: the relevance of association studies in amyotrophic lateral sclerosis. Front Neurosci. 2020;14:603023.
Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–88.
van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6:869–77.
van Es MA, van Vught PW, Blauw HM, Franke L, Saris CG, Van den Bosch L, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40:29–31.
van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–7.
Landers JE, Melki J, Meininger V, Glass JD, van den Berg LH, van Es MA, et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2009;106:9004–9.
Diekstra FP, Saris CG, van Rheenen W, Franke L, Jansen RC, van Es MA, et al. Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLos ONE. 2012;7:e35333.
Iida A, Takahashi A, Kubo M, Saito S, Hosono N, Ohnishi Y, et al. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet. 2011;20:3684–92.
Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P, et al. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat Genet. 2013;45:697–700.
Fogh I, Ratti A, Gellera C, Lin K, Tiloca C, Moskvina V, et al. A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23:2220–31.
Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41.
Tohnai G, Nakamura R, Sone J, Nakatochi M, Yokoi D, Katsuno M, et al. Frequency and characteristics of the TBK1 gene variants in Japanese patients with sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2018;64:158.e15–58e19.
Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N, et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol. 2020;3:526.
Iacoangeli A, Lin T, Al Khleifat A, Jones AR, Opie-Martin S, Coleman JRI, et al. Genome-wide meta-analysis finds the ACSL5-ZDHHC6 locus is associated with ALS and links weight loss to the disease genetics. Cell Rep. 2020;33:108323.
Johnson JO, Chia R, Miller DE, Li R, Kumaran R, Abramzon Y, et al. Association of variants in the SPTLC1 gene with juvenile amyotrophic lateral sclerosis. JAMA Neurol. 2021;78:1236–48.
van Rheenen W, van der Spek RAA, Bakker MK, van Vugt J, Hop PJ, Zwamborn RAJ, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53:1636–48.
Julian TH, Boddy S, Islam M, Kurz J, Whittaker KJ, Moll T, et al. A review of Mendelian randomization in amyotrophic lateral sclerosis. Brain. 2021;145:832–42.
Zhang S, Cooper-Knock J, Weimer AK, Shi M, Moll T, Marshall JNG, et al. Genome-wide identification of the genetic basis of amyotrophic lateral sclerosis. Neuron. 2022;110;992–1008.e11.
van Es MA, Veldink JH, Saris CGJ, Blauw HM, van Vught PWJ, Birve A, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–7.
van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043.
Benyamin B, He J, Zhao QY, Gratten J, Garton F, Leo PJ, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat. Commun. 2017;8:611.
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268.
Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N, et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun. Biol. 2020;3:526.
van Rheenen W, van der Spek RAA, Bakker MK, van Vugt JJFA, Hop PJ, Zwamborn RAJ, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53:1636.
Tan HHG, Westeneng HJ, van der Burgh HK, van Es MA, Bakker LA, van Veenhuijzen K, et al. The distinct traits of the UNC13A polymorphism in amyotrophic lateral sclerosis. Ann Neurol. 2020;88:796–806.
Ma XR, Prudencio M, Koike Y, Vatsavayai SC, Kim G, Harbinski F, et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature. 2022;603:124–30.
Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603:131–7.
Camacho M, Quade B, Trimbuch T, Xu J, Sari L, Rizo J, et al. Control of neurotransmitter release by two distinct membrane-binding faces of the Munc13-1 C1C2B region. Elife. 2021;10:e72030.
Giannoccaro MP, Bartoletti-Stella A, Piras S, Pession A, De Massis P, Oppi F, et al. Multiple variants in families with amyotrophic lateral sclerosis and frontotemporal dementia related to C9orf72 repeat expansion: further observations on their oligogenic nature. J Neurol. 2017;264:1426–33.
Al Khleifat A, Iacoangeli A, van Vugt J, Bowles H, Moisse M, Zwamborn RAJ, et al. Structural variation analysis of 6,500 whole genome sequences in amyotrophic lateral sclerosis. NPJ Genom Med. 2022;7:8.
Baxi EG, Thompson T, Li J, Kaye JA, Lim RG, Wu J, et al. Answer ALS, a large-scale resource for sporadic and familial ALS combining clinical and multi-omics data from induced pluripotent cell lines. Nat Neurosci. 2022;25:226–37.
Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–13.
Pang SY, Hsu JS, Teo KC, Li Y, Kung MHW, Cheah KSE, et al. Burden of rare variants in ALS genes influences survival in familial and sporadic ALS. Neurobiol Aging. 2017;58:238.e9–15.
Naruse H, Ishiura H, Mitsui J, Takahashi Y, Matsukawa T, Tanaka M, et al. Burden of rare variants in causative genes for amyotrophic lateral sclerosis (ALS) accelerates age at onset of ALS. J Neurol Neurosurg Psychiatry. 2019;90:537–42.
Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–75.
Hart MP, Gitler AD. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–42.
Naruse H, Matsukawa T, Ishiura H, Mitsui J, Takahashi Y, Takano H, et al. Association of ATXN2 intermediate-length CAG repeats with amyotrophic lateral sclerosis correlates with the distributions of normal CAG repeat alleles among individual ethnic populations. Neurogenetics. 2019;20:65–71.
Becker LA, Huang B, Bieri G, Ma R.Knowles DA, Jafar-Nejad P, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–71.
Perrone B, Conforti FL. Common mutations of interest in the diagnosis of amyotrophic lateral sclerosis: how common are common mutations in ALS genes? Expert Rev Mol Diagn. 2020;20:703–14.
Shepheard SR, Parker MD, Cooper-Knock J, Verber NS, Tuddenham L, Heath P, et al. Value of systematic genetic screening of patients with amyotrophic lateral sclerosis. J Neurol Neurosur Psychiatry. 2021;92:510–8.
Brown CA, Lally C, Kupelian V, Flanders WD. Estimated prevalence and incidence of amyotrophic lateral sclerosis and SOD1 and C9orf72 genetic variants. Neuroepidemiology. 2021;55:342–53.
Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15.
Grassano M, Calvo A, Moglia C, Brunetti M, Barberis M, Sbaiz L, et al. Mutational analysis of known ALS genes in an Italian population-based cohort. Neurology. 2021;96:E600–9.
Calvo A, Moglia C, Canosa A, Brunetti M, Barberis M, Traynor BJ, et al. A de novo nonsense mutation of the FUS gene in an apparently familial amyotrophic lateral sclerosis case. Neurobiol Aging. 2014;35:1513.e7–11.
Hubers A, Just W, Rosenbohm A, Muller K, Marroquin N, Goebel I, et al. De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiol Aging. 2015;36:3117.e1–e6.
Laffita-Mesa JM, Pupo JMR, Sera RM, Mojena YV, Kouri V, Laguna-Salvia L, et al. De novo mutations in ataxin-2 gene and ALS risk. PLos ONE. 2013;8:e70560.
Takahashi Y, Fukuda Y, Yoshimura J, Toyoda A, Kurppa K, Moritoyo H, et al. ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am J Hum Genet. 2013;93:900–5.
Heo K, Lim SM, Nahm M, Kim YE, Oh KW, Park HT, et al. A de novo RAPGEF2 variant identified in a sporadic amyotrophic lateral sclerosis patient impairs microtubule stability and axonal mitochondria distribution. Exp Neurobiol. 2018;27:550–63.
Salmon K, Kiernan MC, Kim SH, Andersen PM, Chio A, van den Berg LH, et al. The importance of offering early genetic testing in everyone with amyotrophic lateral sclerosis. Brain. 2022;145:1207–10.
Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21.
Amado DA, Davidson BL. Gene therapy for ALS: a review. Mol Ther. 2021;29:3345–58.
Mathis S, Le Masson G. RNA-targeted therapies and amyotrophic lateral sclerosis. Biomedicines. 2018;6:9.
Ly CV, Miller TM. Emerging antisense oligonucleotide and viral therapies for amyotrophic lateral sclerosis. Curr Opin Neurol. 2018;31:648–54.
Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90:535–50.
Tran H, Moazami MP, Yang H, McKenna-Yasek D, Douthwright CL, Pinto C, et al. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat Med. 2021;28:117–4.
Korobeynikov VA, Lyashchenko AK, Blanco-Redondo B, Jafar-Nejad P, Shneider NA. Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat Med. 2022;28:104–16.
Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22:167–79.
Gennemark P, Walter K, Clemmensen N, Rekic D, Nilsson CAM, Knochel J, et al. An oral antisense oligonucleotide for PCSK9 inhibition. Sci Transl Med. 2021;13:eabe9117.
Nagata T, Dwyer CA, Yoshida-Tanaka K, Ihara K, Ohyagi M, Kaburagi H, et al. Cholesterol-functionalized DNA/RNA heteroduplexes cross the blood-brain barrier and knock down genes in the rodent CNS. Nat Biotechnol. 2021;39:1529–36.
Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, Goodspeed K, Gray SJ, Kay CN, et al. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther. 2021;29:464–88.
Duque S, Joussemet B, Riviere C, Marais T, Dubreil L, Douar AM, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–96.
Day JW, Finkel RS, Chiriboga CA, Connolly AM, Crawford TO, Darras BT, et al. Onasemnogene abeparvovec gene therapy for symptomatic infantile-onset spinal muscular atrophy in patients with two copies of SMN2 (STR1VE): an open-label, single-arm, multicentre, phase 3 trial. Lancet Neurol. 2021;20:284–93.
Liu HN, Tjostheim S, Dasilva K, Taylor D, Zhao B, Rakhit R, et al. Targeting of monomer/misfolded SOD1 as a therapeutic strategy for amyotrophic lateral sclerosis. J Neurosci. 2012;32:8791–9.
Tamaki Y, Shodai A, Morimura T, Hikiami R, Minamiyama S, Ayaki T, et al. Elimination of TDP-43 inclusions linked to amyotrophic lateral sclerosis by a misfolding-specific intrabody with dual proteolytic signals. Sci Rep. 2018;8.
Bush JA, Aikawa H, Fuerst R, Li Y, Ursu A, Meyer SM, et al. Ribonuclease recruitment using a small molecule reduced c9ALS/FTD r(G(4)C(2)) repeat expansion in vitro and in vivo ALS models. Sci Transl Med. 2021;13:eabd5991.
Diller DJ, Swanson J, Bayden AS, Jarosinski M, Audie J. Rational, computer-enabled peptide drug design: principles, methods, applications and future directions. Future Med Chem. 2015;7:2173–93.
Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Disco. 2002;1:727–30.
Ichiyanagi N, Fujimori K, Yano M, Ishihara-Fujisaki C, Sone T, Akiyama T, et al. Establishment of in vitro FUS-associated familial amyotrophic lateral sclerosis model using human induced pluripotent stem cells. Stem Cell Rep. 2016;6:496–510.
Okano H, Morimoto S. iPSC-based disease modeling and drug discovery in cardinal neurodegenerative disorders. Cell Stem Cell. 2022;29:189–208.
Imamura K, Izumi Y, Watanabe A, Tsukita K, Woltjen K, Yamamoto T, et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:eaaf3962.
Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med. 2018;24:1579–89.
Morimoto S, Takahashi S, Ito D, Daté Y, Okada K, Muh Chyi C, et al. Ropinirole hydrochloride for amyotrophic lateral sclerosis: a single-center, randomized feasibility, double-blind, placebo-controlled trial. medRxiv 2021.12.05.21267266; https://doi.org/10.1101/2021.12.05.21267266 [Preprint]. 2021.
Wainger BJ, Macklin EA, Vucic S, McIlduff CE, Paganoni S, Maragakis NJ, et al. Effect of ezogabine on cortical and spinal motor neuron excitability in amyotrophic lateral sclerosis: a randomized clinical trial. JAMA Neurol. 2021;78:186–96.
Nagai M, Aoki M, Miyoshi I, Kato M, Pasinelli P, Kasai N, et al. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J Neurosci. 2001;21:9246–54.
Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, et al. Molecular cloning and expression of human hepatocyte growth factor. Nature 1989;342:440–3.
Sun W, Funakoshi H, Nakamura T. Overexpression of HGF retards disease progression and prolongs life span in a transgenic mouse model of ALS. J Neurosci. 2002;22:6537–48.
Ishigaki A, Aoki M, Nagai M, Warita H, Kato S, Kato M, et al. Intrathecal delivery of hepatocyte growth factor from amyotrophic lateral sclerosis onset suppresses disease progression in rat amyotrophic lateral sclerosis model. J Neuropathol Exp Neurol. 2007;66:1037–44.
Brettschneider J, Petzold A, Sussmuth SD, Ludolph AC, Tumani H. Axonal damage markers in cerebrospinal fluid are increased in ALS. Neurology 2006;66:852–6.
Steinacker P, Feneberg E, Weishaupt J, Brettschneider J, Tumani H, Andersen PM, et al. Neurofilaments in the diagnosis of motoneuron diseases: a prospective study on 455 patients. J Neurol Neurosurg Psychiatry. 2016;87:12–20.
Bjornevik K, O’Reilly EJ, Molsberry S, Kolonel LN, Le Marchand L, Paganoni S, et al. Prediagnostic Neurofilament Light Chain Levels in Amyotrophic Lateral Sclerosis. Neurology. 2021.
Paganoni S, Macklin EA, Hendrix S, Berry JD, Elliott MA, Maiser S, et al. Trial of Sodium Phenylbutyrate-Taurursodiol for Amyotrophic Lateral Sclerosis. N. Engl J Med. 2020;383:919–30.
Magen I, Yacovzada NS, Yanowski E, Coenen-Stass A, Grosskreutz J, Lu CH, et al. Circulating miR-181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat Neurosci. 2021;24:1534–41.
Manberg A, Skene N, Sanders F, Trusohamn M, Remnestal J, Szczepinska A, et al. Altered perivascular fibroblast activity precedes ALS disease onset. Nat Med. 2021;27:640–6.
Berry JD, Miller R, Moore DH, Cudkowicz ME, van den Berg LH, Kerr DA, et al. The Combined Assessment of Function and Survival (CAFS): a new endpoint for ALS clinical trials. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14:162–8.
Bozik ME, Mitsumoto H, Brooks BR, Rudnicki SA, Moore DH, Zhang B, et al. A post hoc analysis of subgroup outcomes and creatinine in the phase III clinical trial (EMPOWER) of dexpramipexole in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15:406–13.
Cudkowicz M, Genge A, Maragakis N, Petri S, van den Berg L, Aho VV, et al. Safety and efficacy of oral levosimendan in people with amyotrophic lateral sclerosis (the REFALS study): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Neurol. 2021;20:821–31.
Morimoto S, Takahashi S, Fukushima K, Saya H, Suzuki N, Aoki M, et al. Ropinirole hydrochloride remedy for amyotrophic lateral sclerosis - Protocol for a randomized, double-blind, placebo-controlled, single-center, and open-label continuation phase I/IIa clinical trial (ROPALS trial). Regen Ther. 2019;11:143–66.
van Eijk RPA, van den Berg LH, Lu Y. Composite endpoint for ALS clinical trials based on patient preference: Patient-Ranked Order of Function (PROOF). J Neurol Neurosurg Psychiatry. 2021.
Wong C, Stavrou M, Elliott E, Gregory JM, Leigh N, Pinto AA, et al. Clinical trials in amyotrophic lateral sclerosis: a systematic review and perspective. Brain Commun. 2021;3:fcab242.
Muller K, Oh KW, Nordin A, Panthi S, Kim SH, Nordin F, et al. De novo mutations in SOD1 are a cause of ALS. J Neurol Neurosur Ps. 2021.
Tian F, Yang W, Mordes DA, Wang JY, Salameh JS, Mok J, et al. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat Commun. 2016;7:13283.
Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–40.
Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 2014;84:324–31.
Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen TT, Racher H, et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015;17:1074–87.
Watanabe Y, Nakagawa T, Akiyama T, Nakagawa M, Suzuki N, Warita H, et al. An Amyotrophic Lateral Sclerosis-Associated Mutant of C21ORF2 Is Stabilized by NEK1-Mediated Hyperphosphorylation and the Inability to Bind FBXO3. iScience. 2020;23:101491.
Briese M, Saal L, Appenzeller S, Moradi M, Baluapuri A, Sendtner M. Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 2016;44:e33.
Rotem N, Magen I, Ionescu A, Gershoni-Emek N, Altman T, Costa CJ, et al. ALS Along the Axons - Expression of Coding and Noncoding RNA Differs in Axons of ALS models. Sci Rep. 2017;7:44500.
Suzuki N, Akiyama T, Warita H, Aoki M. Omics Approach to Axonal Dysfunction of Motor Neurons in Amyotrophic Lateral Sclerosis (ALS). Front Neurosci. 2020;14:194.
Kawada J, Kaneda S, Kirihara T, Maroof A, Levi T, Eggan K, et al. Generation of a Motor Nerve Organoid with Human Stem Cell-Derived Neurons. Stem Cell Rep. 2017;9:1441–9.
Okano H, Yamanaka S. iPS cell technologies: significance and applications to CNS regeneration and disease. Mol Brain. 2014;7:22.
Akiyama T, Suzuki N, Ishikawa M, Fujimori T, Sone K,. Aberrant axon branching via Fos-B dysregulation in FUS-ALS motor neurons. EBioMedicine. 2019;(in press).
Lafragette A, Bardo MT, Lardeux V, Solinas M, Thiriet N. Reduction of Cocaine-Induced Locomotor Effects by Enriched Environment Is Associated with Cell-Specific Accumulation of DeltaFosB in Striatal and Cortical Subregions. Int J Neuropsychopharmacol. 2017;20:237–46.
Cahill ME, Walker DM, Gancarz AM, Wang ZJ, Lardner CK, Bagot RC, et al. The dendritic spine morphogenic effects of repeated cocaine use occur through the regulation of serum response factor signaling. Mol Psychiatry. 2018;23:1474–86.
Anastasiadou S, Knoll B. The multiple sclerosis drug fingolimod (FTY720) stimulates neuronal gene expression, axonal growth and regeneration. Exp Neurol. 2016;279:243–60.
Bhinge A, Namboori SC, Zhang X, VanDongen AMJ, Stanton LW. Genetic correction of SOD1 mutant iPSCs reveals ERK and JNK activated AP1 as a driver of neurodegeneration in amyotrophic lateral sclerosis. Stem Cell Rep. 2017;8:856–69.
Mitsuzawa S, Suzuki N, Akiyama T, Ishikawa M, Sone T, Kawada J, et al. Reduced PHOX2B stability causes axonal growth impairment in motor neurons with TARDBP mutations. Stem Cell Rep. 2021;16:1527–41.
Allodi I, Nijssen J, Benitez JA, Schweingruber C, Fuchs A, Bonvicini G, et al. Modeling motor neuron resilience in ALS using stem cells. Stem Cell Rep. 2019;12:1329–41.
Lee H, Lee JJ, Park NY, Dubey SK, Kim T, Ruan K, et al. Multi-omic analysis of selectively vulnerable motor neuron subtypes implicates altered lipid metabolism in ALS. Nat Neurosci 2021;24:1673–85.
Farias J, Holt CE, Sotelo JR, Sotelo-Silveira JR. Axon microdissection and transcriptome profiling reveals the in vivo RNA content of fully differentiated myelinated motor axons. Rna 2020;26:595–612.
Moloney EB, de Winter F, Verhaagen J. ALS as a distal axonopathy: molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Front Neurosci. 2014;8:252.
Ohkawara B, Ito M, Ohno K. Secreted Signaling Molecules at the Neuromuscular Junction in Physiology and Pathology. Int J Mol Sci. 2021;22.
Martineau E, Di Polo A, Vande Velde C, Robitaille R. Dynamic neuromuscular remodeling precedes motor-unit loss in a mouse model of ALS. Elife. 2018;7.
Miyoshi S, Tezuka T, Arimura S, Tomono T, Okada T, Yamanashi Y. DOK7 gene therapy enhances motor activity and life span in ALS model mice. EMBO Mol Med. 2017;9:880–9.
Wang L, Gao J, Liu J, Siedlak SL, Torres S, Fujioka H, et al. Mitofusin 2 Regulates Axonal Transport of Calpastatin to Prevent Neuromuscular Synaptic Elimination in Skeletal Muscles. Cell Metab. 2018;28:400–14.e8.
Altman T, Ionescu A, Ibraheem A, Priesmann D, Gradus-Pery T, Farberov L, et al. Axonal TDP-43 condensates drive neuromuscular junction disruption through inhibition of local synthesis of nuclear encoded mitochondrial proteins. Nat Commun. 2021;12:6914.
Picchiarelli G, Demestre M, Zuko A, Been M, Higelin J, Dieterle S, et al. FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat Neurosci. 2019;22:1793–805.
Schoen M, Reichel JM, Demestre M, Putz S, Deshpande D, Proepper C, et al. Super-Resolution Microscopy Reveals Presynaptic Localization of the ALS/FTD Related Protein FUS in Hippocampal Neurons. Front Cell Neurosci. 2015;9:496.
So E, Mitchell JC, Memmi C, Chennell G, Vizcay-Barrena G, Allison L, et al. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum Mol Genet. 2018;27:463–74.
Armstrong GA, Drapeau P. Loss and gain of FUS function impair neuromuscular synaptic transmission in a genetic model of ALS. Hum Mol Genet. 2013;22:4282–92.
Westergard T, Jensen BK, Wen X, Cai J, Kropf E, Iacovitti L, et al. Cell-to-cell transmission of dipeptide repeat proteins linked to C9orf72-ALS/FTD. Cell Rep. 2016;17:645–52.
Porta S, Xu Y, Restrepo CR, Kwong LK, Zhang B, Brown HJ, et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun. 2018;9:4220.
Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013;4:124–34.
Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–72.
Stoklund Dittlau K, Krasnow EN, Fumagalli L, Vandoorne T, Baatsen P, Kerstens A, et al. Generation of human motor units with functional neuromuscular junctions in microfluidic devices. J Vis Exp. 2021. https://doi.org/10.3791/62959.
Osaki T, Uzel SGM, Kamm RD. On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat Protoc. 2020;15:421–49.
Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53.
Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–98.e7.
Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med. 2019;25:561–68.
Xing L, Bassell GJ. mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic. 2013;14:2–14.
Riva N, Clarelli F, Domi T, Cerri F, Gallia F, Trimarco A, et al. Unraveling gene expression profiles in peripheral motor nerve from amyotrophic lateral sclerosis patients: insights into pathogenesis. Sci Rep. 2016;6:39297.
Verheijen MH, Peviani M, Hendricusdottir R, Bell EM, Lammens M, Smit AB, et al. Increased axonal ribosome numbers is an early event in the pathogenesis of amyotrophic lateral sclerosis. PLoS ONE. 2014;9:e87255.
Nagano S, Jinno J, Abdelhamid RF, Jin Y, Shibata M, Watanabe S, et al. TDP-43 transports ribosomal protein mRNA to regulate axonal local translation in neuronal axons. Acta Neuropathol. 2020;140:695–713.
Rage F, Boulisfane N, Rihan K, Neel H, Gostan T, Bertrand E, et al. Genome-wide identification of mRNAs associated with the protein SMN whose depletion decreases their axonal localization. RNA. 2013;19:1755–66.
Kye MJ, Niederst ED, Wertz MH, Goncalves Ido C, Akten B, Dover KZ, et al. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet. 2014;23:6318–31.
Umegaki Y, Brotons AM, Nakanishi Y, Luo Z, Zhang H, Bonni A, et al. Palladin is a neuron-specific translational target of mTOR signaling that regulates axon morphogenesis. J Neurosci. 2018;38:4985–95.
Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the survival of motor neuron protein impairs mRNA localization and local translation in the growth cone of motor neurons. J Neurosci. 2016;36:3811–20.
Groen EJ, Fumoto K, Blokhuis AM, Engelen-Lee J, Zhou Y, van den Heuvel DM, et al. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet. 2013;22:3690–704.
Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806.
Lopez-Erauskin J, Tadokoro T, Baughn MW, Myers B, McAlonis-Downes M, Chillon-Marinas C, et al. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron. 2018;100:816–30.
Thomas-Jinu S, Gordon PM, Fielding T, Taylor R, Smith BN, Snowden V, et al. Non-nuclear pool of splicing factor SFPQ regulates axonal transcripts required for normal motor development. Neuron. 2017;94:931.
Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19:690–6.
Takeuchi A, Iida K, Tsubota T, Hosokawa M, Denawa M, Brown JB, et al. Loss of Sfpq causes long-gene transcriptopathy in the brain. Cell Rep. 2018;23:1326–41.
Sahoo PK, Kar AN, Samra N, Terenzio M, Patel P, Lee SJ, et al. A Ca(2+)-dependent switch activates axonal casein kinase 2alpha translation and drives G3BP1 granule disassembly for axon regeneration. Curr Biol. 2020;30:4882–95.
Dalla Costa I, Buchanan CN, Zdradzinski MD, Sahoo PK, Smith TP, Thames E, et al. The functional organization of axonal mRNA transport and translation. Nat Rev Neurosci. 2021;22:77–91.
Maday S, Holzbaur EL. Compartment-specific regulation of autophagy in primary neurons. J Neurosci. 2016;36:5933–45.
Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64.
Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun. 2018;9:335.
Nakazawa S, Oikawa D, Ishii R, Ayaki T, Takahashi H, Takeda H, et al. Linear ubiquitination is involved in the pathogenesis of optineurin-associated amyotrophic lateral sclerosis. Nat Commun. 2016;7:12547.
Li F, Xie X, Wang Y, Liu J, Cheng X, Guo Y, et al. Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat Commun. 2016;7:12708.
Ciura S, Sellier C, Campanari ML, Charlet-Berguerand N, Kabashi E. The most prevalent genetic cause of ALS-FTD, C9orf72 synergizes the toxicity of ATXN2 intermediate polyglutamine repeats through the autophagy pathway. Autophagy. 2016;12:1406–8.
Ito H, Fujita K, Nakamura M, Wate R, Kaneko S, Sasaki S, et al. Optineurin is co-localized with FUS in basophilic inclusions of ALS with FUS mutation and in basophilic inclusion body disease. Acta Neuropathol. 2011;121:555–7.
Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015;130:77–92.
Benyamin B, He J, Zhao Q, Gratten J, Garton F, Leo PJ, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun. 2017;8:611.
Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–16.
Hogan AL, Don EK, Rayner SL, Lee A, Laird AS, Watchon M, et al. Expression of ALS/FTD-linked mutant CCNF in zebrafish leads to increased cell death in the spinal cord and an aberrant motor phenotype. Hum Mol Genet. 2017;26:2616–26.
Williams KL, Topp S, Yang S, Smith B, Fifita JA, Warraich ST, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253.
Edens BM, Yan J, Miller N, Deng HX, Siddique T, Ma YC. A novel ALS-associated variant in UBQLN4 regulates motor axon morphogenesis. Elife. 2017;6:e25453.
Morrice JR, Gregory-Evans CY, Shaw CA. Modeling environmentally-induced motor neuron degeneration in zebrafish. Sci Rep. 2018;8:4890.
Chamberlain KA, Sheng ZH. Mechanisms for the maintenance and regulation of axonal energy supply. J Neurosci Res. 2019;97:897–913.
Zhang CL, Rodenkirch L, Schultz JR, Chiu SY. A novel method to study the local mitochondrial fusion in myelinated axons in vivo. J Neurosci Methods. 2012;207:51–8.
Zhou B, Yu P, Lin MY, Sun T, Chen Y, Sheng ZH. Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits. J Cell Biol. 2016;214:103–19.
Heo K, Lim SM, Nahm M, Kim YE, Oh KW, Park HT, et al. A de novo RAPGEF2 variant identified in a sporadic amyotrophic lateral sclerosis patient impairs microtubule stability and axonal mitochondria distribution. Exp Neurobiol. 2018;27:550–63.
Moller A, Bauer CS, Cohen RN, Webster CP, De Vos KJ. Amyotrophic lateral sclerosis-associated mutant SOD1 inhibits anterograde axonal transport of mitochondria by reducing Miro1 levels. Hum Mol Genet. 2017;26:4668–79.
Denton K, Mou Y, Xu CC, Shah D, Chang J, Blackstone C, et al. Impaired mitochondrial dynamics underlie axonal defects in hereditary spastic paraplegias. Hum Mol Genet. 2018;27:2517–30.
Lin MY, Cheng XT, Xie Y, Cai Q, Sheng ZH. Removing dysfunctional mitochondria from axons independent of mitophagy under pathophysiological conditions. Autophagy. 2017;13:1792–4.
Bernard-Marissal N, Medard JJ, Azzedine H, Chrast R. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain. 2015;138:875–90.
Watanabe S, Ilieva H, Tamada H, Nomura H, Komine O, Endo F, et al. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med. 2016;8:1421–37.
Gaweda-Walerych K, Sitek EJ, Narozanska E, Buratti E. Parkin beyond Parkinson’s disease-A functional meaning of Parkin downregulation in TDP-43 proteinopathies. Cells. 2021;10:3389.
Palomo GM, Granatiero V, Kawamata H, Konrad C, Kim M, Arreguin AJ, et al. Parkin is a disease modifier in the mutant SOD1 mouse model of ALS. EMBO Mol Med. 2018;10:e8888.
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–68.
Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–97.
Chen Y, Deng J, Wang P, Yang M, Chen X, Zhu L, et al. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet. 2016;25:5059–68.
Mehta AR, Gregory JM, Dando O, Carter RN, Burr K, Nanda J, et al. Mitochondrial bioenergetic deficits in C9orf72 amyotrophic lateral sclerosis motor neurons cause dysfunctional axonal homeostasis. Acta Neuropathol. 2021;141:257–79.
Xie Y, Zhou B, Lin MY, Wang S, Foust KD, Sheng ZH. Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron. 2015;87:355–70.
Ono Y, Tanaka H, Takata M, Nagahara Y, Noda Y, Tsuruma K, et al. SA4503, a sigma-1 receptor agonist, suppresses motor neuron damage in in vitro and in vivo amyotrophic lateral sclerosis models. Neurosci Lett. 2014;559:174–8.
Smith R, Pioro E, Myers K, Sirdofsky M, Goslin K, Meekins G, et al. Enhanced bulbar function in amyotrophic lateral sclerosis: the nuedexta treatment trial. Neurotherapeutics. 2017;14:762–72.
Matsuhashi T, Sato T, Kanno SI, Suzuki T, Matsuo A, Oba Y, et al. Mitochonic acid 5 (MA-5) facilitates ATP synthase oligomerization and cell survival in various mitochondrial diseases. EBioMedicine. 2017;20:27–38.
De Vos KJ, Hafezparast M. Neurobiology of axonal transport defects in motor neuron diseases: opportunities for translational research? Neurobiol Dis. 2017;105:283–99.
Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–80.
Rouleau GA, Clark AW, Rooke K, Pramatarova A, Krizus A, Suchowersky O, et al. SOD1 mutation is associated with accumulation of neurofilaments in amyotrophic lateral sclerosis. Ann Neurol. 1996;39:128–31.
Hirano A, Donnenfeld H, Sasaki S, Nakano I. Fine structural observations of neurofilamentous changes in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:461–70.
Hirano A, Nakano I, Kurland LT, Mulder DW, Holley PW, Saccomanno G. Fine structural study of neurofibrillary changes in a family with amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:471–80.
Takahashi T, Yagishita S, Amano N, Yamaoka K, Kamei T. Amyotrophic lateral sclerosis with numerous axonal spheroids in the corticospinal tract and massive degeneration of the cortex. Acta Neuropathol. 1997;94:294–9.
De Vos KJ, Chapman AL, Tennant ME, Manser C, Tudor EL, Lau KF, et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum Mol Genet. 2007;16:2720–8.
Williamson TL, Cleveland DW. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci. 1999;2:50–6.
Magrane J, Cortez C, Gan WB, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum Mol Genet. 2014;23:1413–24.
Gibbs KL, Kalmar B, Rhymes ER, Fellows AD, Ahmed M, Whiting P, et al. Inhibiting p38 MAPK alpha rescues axonal retrograde transport defects in a mouse model of ALS. Cell Death Dis. 2018;9:596.
Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–43.
Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. 2017;8:861.
Duncan JE, Goldstein LS. The genetics of axonal transport and axonal transport disorders. PLoS Genet. 2006;2:e124.
Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–6.
Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–6.
Yan S, Guo C, Hou G, Zhang H, Lu X, Williams JC, et al. Atomic-resolution structure of the CAP-Gly domain of dynactin on polymeric microtubules determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2015;112:14611–6.
Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–96.
Fichera M, Lo Giudice M, Falco M, Sturnio M, Amata S, Calabrese O, et al. Evidence of kinesin heavy chain (KIF5A) involvement in pure hereditary spastic paraplegia. Neurology. 2004;63:1108–10.
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268–83.e6.
Brenner D, Yilmaz R, Muller K, Grehl T, Petri S, Meyer T, et al. Hot-spot KIF5A mutations cause familial ALS. Brain. 2018;141:688–97.
Ebbing B, Mann K, Starosta A, Jaud J, Schols L, Schule R, et al. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet. 2008;17:1245–52.
Citterio A, Arnoldi A, Panzeri E, Merlini L, D’Angelo MG, Musumeci O, et al. Variants in KIF1A gene in dominant and sporadic forms of hereditary spastic paraparesis. J Neurol. 2015;262:2684–90.
Ikenaka K, Katsuno M, Kawai K, Ishigaki S, Tanaka F, Sobue G. Disruption of axonal transport in motor neuron diseases. Int J Mol Sci. 2012;13:1225–38.
Nakamura R, Tohnai G, Atsuta N, Nakatochi M, Hayashi N, Watanabe H, et al. Genetic and functional analysis of KIF5A variants in Japanese patients with sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2021;97:147. e11–47 e17.
Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179:147–64.
Smith BN, Topp SD, Fallini C, Shibata H, Chen HJ, Troakes C, et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:eaad9157.
Cioni JM, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, et al. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176:56–72.e15.
Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–76.
Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503.
Fil D, DeLoach A, Yadav S, Alkam D, MacNicol M, Singh A, et al. Mutant Profilin1 transgenic mice recapitulate cardinal features of motor neuron disease. Hum Mol Genet. 2017;26:686–701.
Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci. 2016;19:1610–18.
Moradi M, Sivadasan R, Saal L, Luningschror P, Dombert B, Rathod RJ, et al. Differential roles of alpha-, beta-, and gamma-actin in axon growth and collateral branch formation in motoneurons. J Cell Biol. 2017;216:793–814.
Bergeron C, Beric-Maskarel K, Muntasser S, Weyer L, Somerville MJ, Percy ME. Neurofilament light and polyadenylated mRNA levels are decreased in amyotrophic lateral sclerosis motor neurons. J Neuropathol Exp Neurol. 1994;53:221–30.
Corbo M, Hays AP. Peripherin and neurofilament protein coexist in spinal spheroids of motor neuron disease. J Neuropathol Exp Neurol. 1992;51:531–7.
Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–61.
Melamed Z, Lopez-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci. 2019;22:180–90.
Menon S, Gupton S. Recent advances in branching mechanisms underlying neuronal morphogenesis. F1000Res. 2018;7:F1000.
Osking Z, Ayers JI, Hildebrandt R, Skruber K, Brown H, Ryu D, et al. ALS-linked SOD1 mutants enhance neurite outgrowth and branching in adult motor neurons. iScience. 2019;11:294–304.
Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–83.
Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van den Bosch L, Van Damme P, et al. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS ONE. 2010;5:e13368.
Garone MG, Birsa N, Rosito M, Salaris F, Mochi M, de Turris V, et al. ALS-related FUS mutations alter axon growth in motoneurons and affect HuD/ELAVL4 and FMRP activity. Commun Biol. 2021;4:1025.
McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–31.
Cheng F, De Luca A, Hogan AL, Rayner SL, Davidson JM, Watchon M, et al. Unbiased label-free quantitative proteomics of cells expressing amyotrophic lateral sclerosis (ALS) mutations in CCNF reveals activation of the apoptosis pathway: a workflow to screen pathogenic gene mutations. Front Mol Neurosci. 2021;14:627740.
Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–24.
Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38.
Kim HJ, Taylor JP. Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron. 2017;96:285–97.
Nedelsky NB, Taylor JP. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol. 2019;15:272–86.
Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506.
Bogaert E, Boeynaems S, Kato M, Guo L, Caulfield TR, Steyaert J, et al. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 2018;24:529–37.
Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–27.
Rhoads SN, Monahan ZT, Yee DS, Leung AY, Newcombe CG, O’Meally RN, et al. The prionlike domain of FUS is multiphosphorylated following DNA damage without altering nuclear localization. Mol Biol Cell. 2018;29:1786–97.
Cicardi ME, Marrone L, Azzouz M, Trotti D. Proteostatic imbalance and protein spreading in amyotrophic lateral sclerosis. EMBO J. 2021;40:e106389.
Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173:677–92.
Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018;173:693–705.e22.
Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–19.
Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell. 2018;173:720–34.
Gwon Y, Maxwell BA, Kolaitis RM, Zhang P, Kim HJ, Taylor JP. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science. 2021;372:eabf6548.
Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5:e18413.
Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science. 2021;371:eabb4309.
Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173:958–71.
Vanneste J, Van Den Bosch L. The role of nucleocytoplasmic transport defects in amyotrophic lateral sclerosis. Int J Mol Sci. 2021;22:12175.
Fallini C, Khalil B, Smith CL, Rossoll W. Traffic jam at the nuclear pore: all roads lead to nucleocytoplasmic transport defects in ALS/FTD. Neurobiol Dis. 2020;140:104835.
Schmidt HB, Gorlich D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci. 2016;41:46–61.
Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 2014;159:388–401.
Coyne AN, Baskerville V, Zaepfel BL, Dickson DW, Rigo F, Bennett F, et al. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci Transl Med. 2021;13:eabe1923.
Lin YC, Kumar MS, Ramesh N, Anderson EN, Nguyen AT, Kim B, et al. Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway. Nat Neurosci. 2021;24:1077–88.
Nanaura H, Kawamukai H, Fujiwara A, Uehara T, Aiba Y, Nakanishi M, et al. C9orf72-derived arginine-rich poly-dipeptides impede phase modifiers. Nat Commun. 2021;12:5301.
Dadon-Nachum M, Melamed E, Offen D. The “Dying-Back” phenomenon of motor neurons in ALS. J Mol Neurosci. 2011;43:470–7.
Baker MR. ALS-dying forward, backward or outward? Nat Rev Neurol. 2014;10:660.
Ghasemi M, Brown RH, Jr. Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med. 2018;8:a024125.
Acknowledgements
We want to express our gratitude to all patients with ALS and their families who cooperated in the gene analysis and their doctors. The authors thank R. Izumi, T. Akiyama, S. Mitsuzawa, Y. Watanabe, A. Ohno, J. Kawada, H. Okano, T. Fujii, A. Gitler, K. Eggan, R. Funayama, K. Nakayama, T. Niihori, Y. Aoki, and all members of the Department of Neurology for the excellent discussion. Moreover, we thank N. Shimakura and M. Suzuki for the technical support and Enago for the English language review (www.enago.jp).
Funding
This work is supported by funding from Grant-in-Aid for Scientific Research (C) (21K07411) and Grant-in-Aid for Scientific Research (B) (20H03586) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; the Research Project for Practical Applications of Regenerative Medicine from the Japan Agency for Medical Research and Development (AMED) (Grant number 21bm0804003h0003); and the Takeda Science Foundation, SERIKA FUND, and Mochida Memorial Foundation for Medical and Pharmaceutical Research.
Author information
Authors and Affiliations
Contributions
NS and MA prepared the manuscript. NS and AN prepared the Table. All authors read, revised, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suzuki, N., Nishiyama, A., Warita, H. et al. Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy. J Hum Genet 68, 131–152 (2023). https://doi.org/10.1038/s10038-022-01055-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01055-8
This article is cited by
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Genetic diagnosis and detection rates using C9orf72 repeat expansion and a multi-gene panel in amyotrophic lateral sclerosis
Journal of Neurology (2024)
-
DNA methylome, R-loop and clinical exome profiling of patients with sporadic amyotrophic lateral sclerosis
Scientific Data (2024)
-
Amyotrophic lateral sclerosis: translating genetic discoveries into therapies
Nature Reviews Genetics (2023)