Abstract
Dyslipidemia is a risk factor for cardiovascular diseases and type 2 diabetes. Several adipokines play important roles in modulation of blood lipids. Metrnl is a recently identified adipokine, and adipose Metrnl participates in regulation of blood triglyceride (TG). In this study, we generated Metrnl global, intestine-specific and liver-specific knockout mice, and explored the effects of Metrnl on serum lipid parameters. Global knockout of Metrnl had no effects on serum lipid parameters under normal chow diet, but increased blood TG by 14%, and decreased total cholesterol (TC) by 16% and high density lipoprotein cholesterol (HDL-C) by 24% under high fat diet. Nevertheless, intestine-specific knockout of Metrnl did not alter the serum lipids parameters under normal chow diet or high fat diet. Notably, liver-specific knockout of Metrnl decreased HDL-C by 24%, TC by 20% and low density lipoprotein cholesterol (LDL-C) by 16% without alterations of blood TG and nonesterified fatty acids (NEFA) under high fat diet. But deficiency of Metrnl in liver did not change VLDL secretion and expression of lipid synthetic and metabolic genes. We conclude that tissue-specific Metrnl controls different components of blood lipids. In addition to modulation of blood TG by adipose Metrnl, blood HDL-C is regulated by liver Metrnl.
Similar content being viewed by others
Introduction
Dyslipidemias, characterized as elevated blood total cholesterol (TC), low-density lipoprotein (LDL) cholesterol or triglycerides (TG) with or without significantly reduced high-density lipoprotein (HDL) cholesterol, are major determinants of atherosclerotic cardiovascular diseases and are associated with hypertension, type 2 diabetes, pancreatitis, Alzheimer’s disease, and so forth [1,2,3]. The prevalence of dyslipidemias is reaching epidemic proportions owing to dietary factors and sedentary lifestyles. The regulatory mechanism of blood lipids needs to be further clarified to find new promising treatments for dyslipidemias.
Numerous adipose tissue-derived secreted proteins, known as adipokines, are associated with blood lipids [4, 5] and include visfatin [4], adiponectin [6], leptin [7], chemerin [8], and progranulin [9]. Metrnl is an adipokine we recently identified that is highly expressed in subcutaneous adipose tissue [10]. It has multiple functions, such as promoting neural development [11], browning white adipose tissue [12] and regulating the immune system [13, 14].
Our previous work has shown that adipose Metrnl can improve high-fat diet-induced insulin resistance by remodeling adipose tissue [15]. Adipose Metrnl can also participate in triglyceride metabolism. Deficiency of adipose Metrnl deteriorates high-fat diet-induced hypertriglyceridemia, while overexpression of Metrnl in adipose tissue improves triglyceride tolerance and high-fat diet-induced hypertriglyceridemia [15]. These results imply that Metrnl may play broad roles in lipid metabolism. However, the overall effects of Metrnl on blood lipids, especially clinical lipid parameters, are unclear. In addition, which tissue contributes to Metrnl-regulated alterations of blood lipids also needs to be identified.
In this study, we detected changes in major clinical lipid parameters induced by global knockout of Metrnl and identified the tissues contributing to Metrnl-induced changes in blood clinical lipid parameters with tissue-specific knockout animal models under both a normal chow diet and a high-fat diet.
Materials and methods
Generation of transgenic animal models
Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Second Military Medical University. Alb-Cre mice, Ella-Cre mice and C57BL/6 mice were purchased from Shanghai Research Center for Model Organisms (Shanghai, China).
Metrnl floxed (MetrnlloxP/loxP) mice, Ella-Cre mice and C57BL/6 mice were used to generate a Metrnl global knockout animal model. The targeting construct of MetrnlloxP/loxP was characterized in our previous studies [15]. Briefly, three LoxP sequences were inserted into the Metrnl allele to flank exon 3 and the coding region of exon 4, which could be excised by Cre recombinase. Ella-Cre mice, which were proven to target the expression of Cre recombinase to the stages of mouse oocytes and preimplantation embryos [16], were used to generate Metrnl global knockout mice. The breeding strategy was that MetrnlloxP/loxP mice were crossed with Ella-Cre mice to generate Metrnlwt/koElla-Cre mice, which were crossed with C57BL/6 mice to generate Metrnlwt/ko mice. Then, Metrnlwt/ko mice were inbred to generate Metrnlko/ko mice (Metrnl−/−).
Alb-Cre mice were used to generate Metrnl liver-specific knockout (Liver-Metrnl−/−) mice. Briefly, MetrnlloxP/loxP mice were crossed with Alb-Cre mice to generate MetrnlloxP/wtAlb-Cre mice, which were further crossed with MetrnlloxP/loxP mice to generate MetrnlloxP/loxPAlb-Cre mice (Liver-Metrnl−/− mice). Metrnl intestine-specific knockout (Intestine-Metrnl−/−) mice were generated by crossing MetrnlloxP/loxP mice with Vil-Cre mice as we described elsewhere [17]. The background of all the mice used in the breeding process was C57BL/6.
Genomic DNA extraction and genotyping by polymerase chain reaction (PCR)
Mouse genomic DNA was extracted with a Mouse Tail Genomic DNA kit (CoWin Biosciences, Beijing, China) according to the manufacturer’s instructions. PCR was then performed to amplify the target gene by adding 2 μL genomic DNA with corresponding primers in a 20 μL final reaction mixture. Cre primer sequences were as follows: forward 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and reverse 5′-GTGAAACAGCATTGCTGTCACTT-3′ (yielding an expected PCR product of 100 bp). β-actin was used as an internal control, and the primer sequences were as follows: forward 5′-CTAGGCCACAGAATTGAAAGATCT-3′ and reverse 5′-GTAGGTGGAAATTCTAGCATCATCC-3′ (324 bp). Wild-type Metrnl gene primer sequences were as follows: forward 5′-GGATGTTTTCTGAGGGTTGGAGGC-3′ and reverse 5′-TTGGCTTTTGGATGAGCGTTTGAG-3′ (289 bp). Floxed (loxP-flanked) Metrnl allele primer sequences were as follows: forward 5′-TGAGGGTTGGAGGCTCCTAGC-3′ and reverse 5′-GGATGAGCGTTTGAGCACAGC-3′ (243 bp). The knockout Metrnl allele primer sequences were as follows: forward 5′-TCCGCTTGTGGTTTCTGTTCA-3′ and reverse 5′-CAGCAGTTCCAATGGGTCAGG-3′ (150 bp).
Animal feeding and treatment
Mice were housed in a standard SPF animal room with the appropriate temperature (23 ~ 25 °C) and lighting (12 h light-dark cycle) conditions. During the feeding period, mice were provided adequate water and a standard chow diet or high-fat diet (HFD) containing approximately 60% of calories from lipids (Research Diets, New Brunswick, NJ, USA). At the end of treatment, mice were fasted for 12 h and sacrificed by an overdose of pentobarbital sodium (100 mg/kg, i.p., Bio-Light, Shanghai, China), venous blood was collected from the inferior vena cava, and tissue samples were carefully excised.
Real-time PCR
Real-time PCR was performed as reported [18]. Briefly, RNA was separated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the instructions of the manufacturer. Two micrograms of RNA was reverse transcribed to cDNA using the M-MLV enzyme (Promega, Madison, WI, USA). Real-time PCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with 1 μL cDNA template in a 20 μL final reaction mixture (95 °C for 15 min; 95 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s, 40 cycles). The relative expression of Metrnl was normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers used in real-time PCR are listed in Table 1.
Detection of mouse serum Metrnl
Mouse serum Metrnl levels were measured by ELISA kits (R&D Systems, Minneapolis, MN, USA), as we described elsewhere [19]. Briefly, capture antibody was incubated in 96-well plates at room temperature overnight. Then, the plates were washed with washing buffer three times and blocked for a minimum of 1 h by adding 300 μL reagent diluent. After further washes, 100 μL serum or standards were added to the plates and incubated for 2 h. Next, 100 μL detection antibody was added to the plates, which were incubated for 2 h after three washings. Subsequently, 100 μL streptavidin-HRP was added, and the plates were incubated for 20 min in the dark. After further washes, 100 μL substrate solution was added, and the plate was incubated for 20 min in the dark, followed by the addition of 50 μL stop solution to stop the reaction. Finally, the optical density was detected by using a microplate reader (Tecan, Grödig, Austria) with a detection wavelength of 450 nm and a reference wavelength of 540 nm.
Detection of serum lipid parameters
Serum samples were sent to Servicebio (Wuhan, Hubei, China), and the clinical serum lipid parameters, including TC, HDL cholesterol, LDL cholesterol, TG and NEFA, were measured by biochemical analyzer (Hitachi, Tokyo, Japan) with the corresponding reagent (Huili Biology, Changchun, China) [20]. Briefly, to measure HDL cholesterol, heparin and MnCl2 were needed to precipitate apoB-containing lipoproteins, and HDL cholesterol was measured from the supernatant. For LDL cholesterol, phosphotungstic acid magnesium and polyethylene glycol were used to precipitate and disperse LDL lipoproteins, and the LDL cholesterol concentration was measured by directly detecting serum absorbance. Considering the high level of free serum glycerol, glycerol concentrations were also determined to correct the triglyceride values. Serum nonesterified fatty acids were determined using commercial assay kits according to the kit instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Cell culture and viral transduction
The isolation and culture of primary hepatocytes were carried out as reported in previous studies [21]. Briefly, primary hepatocytes were isolated from C57/BL6 mice aged 9 weeks by using the collagenase perfusion method. Then, the cells were cultured in plates coated with rat tail collagen in DMEM (HyClone, Logan, UT, USA) with 10% fetal bovine serum and 1% penicillin streptomycin. After 4 h of attachment in a 37 °C incubator chamber (Thermo Scientific, Waltham, MA, USA) with 5% CO2, viruses encoding the Metrnl open reading frame (HANBIO, Shanghai, China) were added to the culture medium at an MOI of 2. After 24 h of infection, primary hepatocytes were changed to fresh medium and cultured in an incubator chamber.
In vivo VLDL secretion assay
In vivo VLDL secretion was detected as described elsewhere [22]. Briefly, after fasting for 16 h, mice were injected with the lipase inhibitor tyloxapol (500 mg/kg, Sigma-Aldrich, St. Louis, MO, USA) through the tail vein. Blood was collected at 0, 1, 2, and 3 h after injection and used for TG measurements with the corresponding kit (APPLYGEN, Beijing, China).
Statistical analysis
All data are expressed as the mean ± SEM. Statistical analysis was performed with Prism 5.0 software. A two-tailed Student’s t-test was used to evaluate the differences. P < 0.05 denoted the presence of a statistically significant difference.
Results
Blood Metrnl levels were elevated in mice fed a high-fat diet
To evaluate the effect of a chronic high-fat diet on circulating Metrnl, C57/BL6 mice were fed a high-fat diet for 7 weeks, and serum Metrnl was assayed with an ELISA. As shown in Fig. 1a, blood Metrnl was significantly increased in mice fed a high-fat diet. In our previous work, we showed that Metrnl expression was upregulated in white adipose tissue in obese mice [10]. Here, we further assessed the influence of a chronic high-fat diet on the expression of Metrnl in the liver and intestine, which showed that Metrnl expression was unchanged in either the liver or intestine under a chronic high-fat diet (Fig. 1b, c).
The effects of chronic high-fat diet on blood Metrnl and expression of Metrnl in liver and intestine. a Chronic high-fat diet increased blood Metrnl levels (n = 3). b, c Metrnl expression in the liver (b) and intestine (c) of mice fed a normal chow diet (NCD) and a high-fat diet (HFD) was detected with real-time PCR (n = 3). *P < 0.05 vs. NCD
Generation of the Metrnl global knockout animal model
To explore the overall function of Metrnl on blood lipids, Metrnl global knockout mice were generated by excision of exon 3 and the coding region of exon 4 of the Metrnl gene, which was proven by genotyping (Fig. 2a, b). The knockout efficiency was verified by real-time PCR with primers targeting the excised regions of the Metrnl open reading frame, and the results showed that Metrnl mRNA was undetectable in the tested tissues, including the liver, intestine, subcutaneous white adipose tissue (WAT) and brown adipose tissue (BAT), in Metrnl−/− mice (Fig. 2c). Consistently, serum Metrnl was detected in each wild-type mouse but was undetected in all Metrnl−/− mice (Fig. 2d). These results suggest that the Metrnl global knockout mouse model was successfully generated. No visible differences were observed between Metrnl−/− and wild-type mice. There were no significant differences in body weight (Fig. 2e).
The generation of Metrnl global knockout mice (Metrnl−/−) and the effects of global knockout of Metrnl on blood lipid parameters under a normal chow diet. a Knockout strategy (left) and breeding strategy (right) for Metrnl−/−. b Genotyping of the Metrnl−/− mice. 1, Metrnlwt/koElla-Cre; 2, Metrnlwt/ko; 3, Metrnlwt/wt; 4, Metrnlko/ko (Metrnl−/−). c Metrnl levels in different tissues between Metrnl−/− and wild-type mice (WT) (n = 6). ***P < 0.001 vs. WT. d Serum Metrnl concentration between Metrnl−/− and WT mice (n = 6). e Body weights of Metrnl−/− and WT mice fed a normal chow diet (n = 8). f The effects of global knockout of Metrnl on high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), total triglyceride (TG) and nonesterified fatty acid (NEFA) in circulation (n = 8)
Global knockout of Metrnl upregulates serum triglyceride and downregulates HDL cholesterol and total cholesterol
Further, the major clinical lipid parameters, including HDL cholesterol, LDL cholesterol, TC, TG, and NEFA, were detected in Metrnl−/− mice fed both normal chow and a high-fat diet. No differences were observed in any of the clinical lipid parameters between Metrnl−/− and wild-type mice fed a normal chow diet (Fig. 2f). However, mice fed a high-fat diet for 16 weeks showed decreased serum HDL cholesterol by 24% (P < 0.01) and total cholesterol by 16% (P < 0.05) and increased serum total triglyceride by 14% (P < 0.05) with LDL cholesterol, NEFA, and body weight unchanged after global Metrnl knockout compared with wild-type mice (Fig. 3a–f).
The effects of global knockout of Metrnl on blood lipid parameters under a high-fat diet (HFD). a Body weights of Metrnl−/− and wild-type mice (WT) fed a HFD (n = 8-9). b–f The effects of global knockout of Metrnl on high-density lipoprotein cholesterol (HDL-C, b), low-density lipoprotein cholesterol (LDL-C, c), total cholesterol (TC, d), total triglyceride (TG, e) and nonesterified fatty acid (NEFA, f) in circulation (n = 8–9). The mice were fed a HFD for 16 weeks. *P < 0.05; **P < 0.01 vs. WT
Deficiency of intestinal Metrnl does not change the blood clinical lipid parameters
Considering that Metrnl is abundantly expressed in the intestinal epithelium [17] and that the intestinal epithelium plays a critical role in the modulation of blood lipids by regulating lipid absorption and transport [23], we speculated that the intestine may contribute to changes in blood lipid parameters after global knockout of Metrnl. To clarify the effects of intestinal Metrnl on blood lipid parameters, Metrnl intestine-specific knockout mice, which were generated and verified as we described elsewhere [17], were employed. Consistent with the results of Metrnl global knockout mice on a normal chow diet, intestine-specific knockout of Metrnl did not change the body weight or clinical lipid parameters under a normal chow diet (Fig. 4a, b). Notably, body weight and clinical lipid parameters, including HDL cholesterol, total cholesterol, and total triglycerides, were not altered under a high-fat diet for 16 weeks in intestinal Metrnl-specific knockout mice compared with their controls (Fig. 4c, d). These results suggest that intestinal Metrnl does not participate in Metrnl-regulated alterations in clinical lipid parameters.
The effects of intestine-specific knockout of Metrnl on blood lipid parameters. a Body weights of intestine-specific Metrnl knockout (Intestine-Metrnl−/−) and wild-type mice (WT) fed a normal chow diet (n = 7–9). b The effects of intestine-specific knockout of Metrnl on blood lipid parameters on a normal chow diet (n = 7–9). c Body weights of Intestine-Metrnl−/− and WT mice fed a high-fat diet (HFD) for 16 weeks. d The effects of intestine-specific knockout of Metrnl on blood lipid parameters under a HFD for 16 weeks (n = 13–14). HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; NEFA, nonesterified fatty acid
Deficiency of liver Metrnl downregulates serum HDL cholesterol, total cholesterol and LDL cholesterol but does not alter total triglycerides
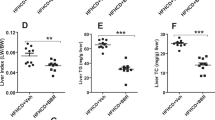
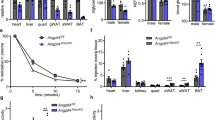
The liver is an important organ involved in the regulation of cholesterol and triglyceride metabolism. To further evaluate the role of liver Metrnl in clinical lipid parameters, Metrnl liver-specific knockout mice (Liver-Metrnl−/−) were first generated by breeding Metrnlloxp/loxp with Alb-Cre (Fig. 5a, b) and confirmed with real-time PCR. Among the detected tissues, including the liver, intestine, WAT and BAT, Metrnl expression in the liver was specifically decreased by approximately 75%, while the expression in other tissues was unchanged (Fig. 5c). Further, the detection of serum Metrnl showed that liver Metrnl-specific knockout did not affect serum Metrnl levels under a normal chow diet (Fig. 5d) or a high-fat diet (Fig. 5e), suggesting that the liver was not an important contributor to blood Metrnl under normal conditions and did not contribute to the obesity-induced increase in blood Metrnl. In line with the results of Metrnl global or intestine-specific knockout mice, no alterations were observed in body weight or clinical lipid parameters under a normal chow diet in Metrnl liver-specific knockout mice (Fig. 5f, g). However, under a high-fat diet for 16 weeks, deficiency of liver Metrnl decreased HDL cholesterol by 24% (P < 0.01), LDL cholesterol by 16% (P < 0.01), and total cholesterol by 20% (P < 0.01), while total triglyceride, nonesterified fatty acids and body weight were unchanged (Fig. 6a–f).
The generation and identification of Metrnl liver-specific knockout mice (Liver-Metrnl−/−) and the effects of liver-specific knockout of Metrnl on blood lipid parameters. a Breeding strategy for Liver-Metrnl−/−. b Genotyping of Liver-Metrnl−/−. 1-3: Liver-Metrnl−/−; 4-6: Metrnlloxp/loxp used as control. c Metrnl levels were detected with real-time PCR in various tissues from Liver-Metrnl−/− and control mice (WT) (n = 6). ***P < 0.001 vs. WT. d Serum Metrnl levels were assayed with ELISA between Liver-Metrnl−/− and WT mice fed a normal chow diet (n = 6). e Serum Metrnl levels were assayed with ELISA between Liver-Metrnl−/− and WT mice fed a high-fat diet (HFD) (n = 3). f Body weights of Liver-Metrnl−/− and WT mice fed a normal chow diet (n = 9–10). g The effects of liver-specific knockout of Metrnl on blood high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), total triglyceride (TG) and nonesterified fatty acid (NEFA) under a normal chow diet (n = 9-10)
The effects of liver-specific knockout of Metrnl on blood lipid parameters under a high-fat diet (HFD). a Body weights of Liver-Metrnl−/− and control mice (WT) fed a HFD for 16 weeks (n = 11). b–f The effects of liver-specific knockout of Metrnl on blood high-density lipoprotein cholesterol (HDL-C, b), low-density lipoprotein cholesterol (LDL-C, c), total cholesterol (TC, d), total triglyceride (TG, e) and nonesterified fatty acid (NEFA, f) levels (n = 11). The mice were fed a HFD for 16 weeks. **P < 0.01 vs. WT
Deficiency of liver Metrnl did not decrease VLDL release
To explore the mechanism of decreased LDL cholesterol in Metrnl liver-specific knockout mice, VLDL release was evaluated by detecting the increase in VLDL-TG after inhibition of lipase using tyloxapol in Metrnl liver-specific knockout mice fed normal chow or a high-fat diet. The results showed that the release of VLDL was not changed in liver-specific knockout mice fed either normal chow or a high-fat diet, suggesting that Metrnl deficiency in the liver did not decrease LDL cholesterol by reducing VLDL release (Fig. 7a, b).
The effects of deficiency of liver Metrnl on VLDL release and expression of lipid synthetic and metabolic genes. a, b VLDL release was evaluated in Metrnl liver-specific knockout mice fed a normal chow diet (a) (n = 3) or a high-fat diet (HFD, b) (n = 3). c Expression of lipid synthesis and metabolic genes in Metrnl-deficient livers (n = 7). d Expression of lipid synthesis and metabolic genes in primary hepatocytes overexpressing Metrnl (Metrnl OE, n = 3–6)
Further, we detected the expression of synthetic and metabolic genes both in liver tissues from Metrnl liver-specific knockout mice fed a high-fat diet and in cultured primary hepatocytes with or without Metrnl overexpression. However, none of the detected genes was downregulated (Fig. 7c, d), suggesting that Metrnl did not regulate blood lipids by changing the expression of synthetic and metabolic genes directly.
Discussion
In this study, we investigated the effects of Metrnl on clinical lipid parameters. A chronic high-fat diet increased circulating Metrnl through adipose tissue but not the liver or intestine. Global knockout of Metrnl did not alter any of the clinical lipid parameters under a normal chow diet but increased blood triglyceride levels and decreased blood total cholesterol and HDL cholesterol levels under a high-fat diet. Intestine-specific knockout of Metrnl exhibited no effects on the main clinical lipid parameters under both a normal chow diet and a high-fat diet. However, liver-specific knockout of Metrnl decreased blood total cholesterol, HDL cholesterol and LDL cholesterol under a high-fat diet.
Metrnl regulates blood triglycerides at least partly through adipose tissue
Global knockout of Metrnl increases blood total triglyceride levels under a high-fat diet. We further explored which tissue contributes to Metrnl-regulated blood triglycerides. Intestine-specific deficiency of Metrnl does not change the blood triglyceride concentration under a normal chow or high-fat diet, suggesting that the intestine does not participate in the regulatory effect of Metrnl on blood triglycerides. Although the liver plays a critical role in triglyceride metabolism, it does not mediate Metrnl-regulated alterations of blood triglycerides either because liver-specific knockout of Metrnl does not affect blood triglycerides under a normal chow or high-fat diet.
However, in our previous work, we observed that adipose tissue-specific knockout of Metrnl increased blood triglyceride levels under a high-fat diet and that adipose-specific overexpression of Metrnl decreased high-fat diet-induced hypertriglyceridemia and improved triglyceride tolerance [15]. These results suggest that the regulatory function of Metrnl in blood triglycerides is at least partly mediated by adipose tissue rather than the liver or intestine.
Metrnl regulates blood cholesterol, especially HDL cholesterol
Apart from blood triglycerides, global knockout of Metrnl can affect total cholesterol and HDL cholesterol. Notably, the total deficiency of Metrnl reduces HDL cholesterol to a much greater extent than total cholesterol (24% vs. 16%), with LDL cholesterol unaltered. Considering that total cholesterol contains HDL cholesterol, it is reasonable to speculate that Metrnl deficiency mainly affects HDL cholesterol rather than total cholesterol. It is well documented that HDL cholesterol has beneficial effects [24], whereas LDL cholesterol is considered “bad” cholesterol [25]. Hence, Metrnl may play a favorable role in the modulation of blood lipids and could be a new target that can both reduce blood triglycerides and increase HDL cholesterol.
The liver contributes to the regulatory effects of Metrnl on blood cholesterol
We wondered which tissue mediates Metrnl-regulated alterations in HDL cholesterol and total cholesterol. Intestine-specific knockout of Metrnl does not influence HDL cholesterol or total cholesterol. Moreover, adipose-specific knockout of Metrnl does not alter blood LDL cholesterol, HDL cholesterol or total cholesterol after a high-fat diet for 8 weeks [15]. However, liver-specific knockout of Metrnl significantly reduced HDL cholesterol and total cholesterol. Consistent with the results of Metrnl−/− mice, liver Metrnl deficiency decreased HDL cholesterol much more than it did total cholesterol. Interestingly, deficiency of liver Metrnl caused a similar decrease in HDL cholesterol that a deficiency of total Metrnl did (both is 24%). These results suggest that the liver contributes to Metrnl total deficiency-induced decreases in HDL cholesterol and total cholesterol. In addition, circulating Metrnl in liver-specific knockout mice was unchanged compared with that in wild-type mice fed a high-fat diet, suggesting that the regulatory function of liver Metrnl on lipid metabolism was not mediated by its endocrine action but by its local effect.
LDL cholesterol was also decreased by knockout of liver Metrnl, which was not observed in global knockout of Metrnl, implying that certain tissues (or some other tissues) may compensate for the decrease in LDL cholesterol induced by liver Metrnl deficiency. Thus, further work should identify other tissues that contribute to the regulatory effect of Metrnl on blood lipids.
The molecular mechanisms by which Metrnl regulates blood lipids in different tissues are still unclear. Our work showed that liver Metrnl did not alter blood lipids by regulating VLDL release or changing the expression of any lipid synthesis and metabolic genes examined, indicating that liver Metrnl might regulate blood lipids by eliminating lipoprotein cholesterol. Further work needs to be done to clarify the exact mechanism of Metrnl in the regulation of lipid metabolism.
In conclusion, total Metrnl deficiency mainly reduces blood HDL cholesterol and increases blood triglycerides. The liver contributes to Metrnl deficiency-induced downregulation of HDL cholesterol, while adipose tissue contributes to the Metrnl deficiency-induced decrease in blood triglycerides. Therefore, Metrnl protein could have therapeutic effects on both hypertriglyceridemia and hypo-high-density lipoproteinemia via different tissues.
References
Genest J, McPherson R, Frohlich J, Anderson T, Campbell N, Carpentier A, et al. 2009 Canadian Cardiovascular Society/Canadian guidelines for the diagnosis and treatment of dyslipidemia and prevention of cardiovascular disease in the adult - 2009 recommendations. Can J Cardiol. 2009;25:567–79.
Goldberg IJ. Clinical review 124: Diabetic dyslipidemia: causes and consequences. J Clin Endocrinol Metab. 2001;86:965–71.
Reitz C. Dyslipidemia and dementia: current epidemiology, genetic evidence, and mechanisms behind the associations. J Alzheimers Dis 2012;30(Suppl 2):S127–45.
Chen Z, Zhao GH, Zhang YK, Shen GS, Xu YJ, Xu NW. Research on the correlation of diabetes mellitus complicated with osteoporosis with lipid metabolism, adipokines and inflammatory factors and its regression analysis. Eur Rev Med Pharmacol Sci. 2017;21:3900–5.
Rahimlou M, Mirzaei K, Keshavarz SA, Hossein-Nezhad A. Association of circulating adipokines with metabolic dyslipidemia in obese versus non-obese individuals. Diabetes Metab Syndr. 2016;10:S60–5.
Coimbra S, Reis F. The protective role of adiponectin for lipoproteins in end-stage renal disease patients: relationship with diabetes and body mass index. Oxid Med Cell Longev. 2019;2019:3021785.
Katsiki N, Mikhailidis DP, Banach M. Leptin, cardiovascular diseases and type 2 diabetes mellitus. Acta Pharmacol Sin. 2018;39:1176–88.
Habib SS, Eshki A, AlTassan B, Fatani D, Helmi H, AlSaif S. Relationship of serum novel adipokine chemerin levels with body composition, insulin resistance, dyslipidemia and diabesity in Saudi women. Eur Rev Med Pharmacol Sci. 2017;21:1296–302.
Korolczuk A, Beltowski J. Progranulin, a new adipokine at the crossroads of metabolic syndrome, diabetes, dyslipidemia and hypertension. Curr Pharm Des. 2017;23:1533–9.
Li ZY, Zheng SL, Wang P, Xu TY, Guan YF, Zhang YJ, et al. Subfatin is a novel adipokine and unlike Meteorin in adipose and brain expression. CNS Neurosci Ther. 2014;20:344–54.
Jorgensen JR, Fransson A, Fjord-Larsen L, Thompson LH, Houchins JP, Andrade N, et al. Cometin is a novel neurotrophic factor that promotes neurite outgrowth and neuroblast migration in vitro and supports survival of spiral ganglion neurons in vivo. Exp Neurol 2012;233:172–81.
Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell 2014;157:1279–91.
Ushach I, Arrevillaga-Boni G, Heller GN, Pone E, Hernandez-Ruiz M, Catalan-Dibene J, et al. Meteorin-like/meteorin-beta is a novel immunoregulatory cytokine associated with inflammation. J Immunol 2018;201:3669–76.
Zheng SL, Li ZY, Song J, Liu JM, Miao CY. Metrnl: a secreted protein with new emerging functions. Acta Pharmacol Sin. 2016;37:571–9.
Li ZY, Song J, Zheng SL, Fan MB, Guan YF, Qu Y, et al. Adipocyte metrnl antagonizes insulin resistance through PPARgamma signaling. Diabetes 2015;64:4011–22.
Dooley TP, Miranda M, Jones NC, DePamphilis ML. Transactivation of the adenovirus EIIa promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development. 1989;107:945–56.
Li ZY, Fan MB, Zhang SL, Qu Y, Zheng SL, Song J, et al. Intestinal Metrnl released into the gut lumen acts as a local regulator for gut antimicrobial peptides. Acta Pharmacol Sin. 2016;37:1458–66.
Wan JF, Chu SF, Zhou X, Li YT, He WB, Tan F, et al. Ursodeoxycholic acid protects interstitial Cajal-like cells in the gallbladder from undergoing apoptosis by inhibiting TNF-alpha expression. Acta Pharmacol Sin. 2018;39:1493–500.
Zheng SL, Li ZY, Zhang Z, Wang DS, Xu J, Miao CY. Evaluation of two commercial enzyme-linked immunosorbent assay kits for the detection of human circulating metrnl. Chem Pharm Bull. 2018;66:391–8.
Ge MX, Niu WX, Ren JF, Cai SY, Yu DK, Liu HT, et al. A novel ASBT inhibitor, IMB17-15, repressed nonalcoholic fatty liver disease development in high-fat diet-fed Syrian golden hamsters. Acta Pharmacol Sin. 2019;40:895–907.
Wang X, Xu TY, Liu XZ, Zhang SL, Wang P, Li ZY, et al. Discovery of novel inhibitors and fluorescent probe targeting NAMPT. Sci Rep. 2015;5:12657.
Mobin MB, Gerstberger S, Teupser D, Campana B, Charisse K, Heim MH, et al. The RNA-binding protein vigilin regulates VLDL secretion through modulation of Apob mRNA translation. Nat Commun 2016;7:12848.
Hussain MM. Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipido. 2014;25:200–6.
Curb JD, Abbott RD, Rodriguez BL, Masaki K, Chen R, Sharp DS, et al. A prospective study of HDL-C and cholesteryl ester transfer protein gene mutations and the risk of coronary heart disease in the elderly. J Lipid Res. 2004;45:948–53.
Brown MS, Goldstein JL. How LDL receptors influence cholesterol and atherosclerosis. Sci Am 1984;251:58–66.
Acknowledgements
This work was supported by grants from the Shanghai Project (16JC1405100), the National Natural Science Foundation of China (81730098 and 81130061) and Medical Innovation Project (16QNP087). This work was also supported by the Open Project Program of the CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
Study conception and design: CYM, ZYL; acquisition of data: QQ, WJH, Si-li Zheng, Sai-long Zhang; analysis and interpretation of data: YYL, ZYL, CYM; drafting of manuscript: ZYL, QQ; critical revision: CYM.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Qi, Q., Hu, Wj., Zheng, Sl. et al. Metrnl deficiency decreases blood HDL cholesterol and increases blood triglyceride. Acta Pharmacol Sin 41, 1568–1575 (2020). https://doi.org/10.1038/s41401-020-0368-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-0368-8
Keywords
This article is cited by
-
Chronic vascular pathogenesis results in the reduced serum Metrnl levels in ischemic stroke patients
Acta Pharmacologica Sinica (2024)
-
Metrnl deficiency retards skin wound healing in mice by inhibiting AKT/eNOS signaling and angiogenesis
Acta Pharmacologica Sinica (2023)
-
Metrnl regulates cognitive dysfunction and hippocampal BDNF levels in D-galactose-induced aging mice
Acta Pharmacologica Sinica (2023)
-
Role of circulating molecules in age-related cardiovascular and metabolic disorders
Inflammation and Regeneration (2022)
-
Allisartan isoproxil reduces mortality of stroke-prone rats and protects against cerebrovascular, cardiac, and aortic damage
Acta Pharmacologica Sinica (2021)