Abstract
Type strains of 72 validated Nocardia species were phylogenetically analyzed based on the multilocus sequence analysis (MLSA) concatenated atpD–groL1–groL2–recA–rpoA–secY–sodA–ychF. Furthermore, their similarity based on digital DNA–DNA hybridization (dDDH) was calculated. Nocardia soli, Nocardia cummidelens and Nocardia salmonicida, Nocardia nova and Nocardia elegans, Nocardia exalbida and Nocardia gamkensis, and Nocardia coubleae and Nocardia ignorata formed coherent clades, respectively. Moreover, each set showed over 70% relatedness by dDDH and shared common phenotypic characteristics. Therefore, we propose a reclassification of Nocardia soli and Nocardia cummidelens as a later heterotypic synonym of Nocardia salmonicida, Nocardia elegans as a later heterotypic synonym of Nocardia nova, Nocardia gamkensis as a later heterotypic synonym of Nocardia exalbida, and Nocardia coubleae as a later heterotypic synonym of Nocardia ignorata.
Similar content being viewed by others
Introduction
The genus Nocardia, first proposed by Trevisan [1], is a member of the family Nocardiaceae, suborder Corynebacterineae [2]. At the time of writing, the genus contains over 110 species with validly published names (http://www.bacterio.net/nocardia.html). The generic, medical, and industrial properties of the genus Nocardia has been reviewed by Goodfellow and Maldonado [3] in detail. Members of the genus are aerobic, Gram-stain positive, weakly acid-fast, non-motile, and mycolic acid-containing actinomycetes that form extensively branched mycelia and substrate hyphae that fragment into rod-shaped, non-motile elements. The genus is characterized chemotaxonomically by the presence of meso-diaminopimelic acid in the cell wall peptidoglycan, arabinose, and galactose as the characteristic sugars in the whole-cell hydrolysates (type IV), diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides as diagnostic phospholipids (type PII), MK-8(H 4,ω-cycl) as the predominant menaquinone, straight-chain and unsaturated fatty acids and tuberculostearic acid as the major cellular fatty acids, and mycolic acid. Most species of the genus were isolated from soil sample, and some of them have been shown to be agents of human and animal diseases. Some Nocardia strains are known as producers of secondary metabolites with diverse biological activities and complex structures such as siderophores, polyketides, and terpenoids. Clarification of the taxonomic relationships among the members of this genus is important for clinical analysis and industrial use. Some molecular approaches have been applied to the identification of Nocardia strains [4,5,6,7]. The analysis of the 16S ribosomal RNA (rRNA) gene sequence still represents the backbone of taxonomic studies, but in some taxa, such as the class Actinobacteria, this gene is too conserved to distinguish two closely related species in many cases [8, 9]. The multilocus sequence analysis (MLSA) was recommended as a genetic method for species definition [10], and is generally used using various sets of genes for the definition of novel species and reclassification of the class Actinobacteria [11,12,13,14,15,16]. Recent studies demonstrated that genome-based methods are able to provide a conceptual framework for bacterial taxonomy of particular species. These methods were also shown to be a digital DNA–DNA hybridization (dDDH) replacement for laboratory DNA–DNA hybridization (DDH), which is needed to describe new species [8, 17,18,19,20,21,22,23,24]. Approaches such as these have proven to be successful and are being increasingly adopted for the definition of novel species of the class Actinobacteria [25,26,27,28]. Previously, we investigated the taxonomic relationships of 26 Nocardia species based on MLSA and dDDH. We determined that the results obtained using these five methods correlated well with each other [29]. In this study, we show the phylogenetic relationships among the 72 Nocardia species analyzed by MLSA, and the reclassification of some Nocardia species based on whole genome sequence and associated phenotypic data.
Materials and methods
The strains of 72 validly proposed Nocardia species preserved at the Biological Resource Center, National Institute of Technology and Evaluation (NBRC), were used in this study (Table 1). DNA extraction was carried out as previously described [29]. Whole genome shotgun sequencing experiments were performed using the next-generation sequencing technique (Illumina MiSeq). The sequences were assembled using Newbler version 2.6 software and subsequently assessed using GenoFinisher software [30, 31]. The DNA sequences of genes encoding adenosine triphosphate (ATP) synthase subunit beta (atpD), chaperonin GroEL (two types of groL), DNA recombination and repair protein (recA), DNA-directed RNA polymerase subunit alpha (rpoA), preprotein translocase subunit SecY (secY), superoxide dismutase (sodA), and ribosome-binding ATPase (ychF) were extracted from each genome sequence. They were then concatenated as pseudo single sequences. Those concatenated sequences were used to perform a similarity search and phylogenetic analysis based on neighbor-joining (NJ), maximum-likelihood (ML), and maximum-parsimony (MP) algorithms using MEGA6 [32]. Genome-to-genome distance (GGD) [33] was computed on whole genome sequences to measure the genetic and evolutionary relatedness among strains, and to help consolidate the existing taxonomic ranks of bacterial strains. The GGD calculations were performed using the Genome-to-genome distance calculator, version 2 (available at http://ggdc.dsmz.de/), and expressed as a percent dDDH. Laboratorial DDH relatedness were determined by the microplate hybridization method developed by Ezaki et al. [34], using five replications. After the highest and lowest values for each sample were excluded, the mean was reported as the DDH relatedness. API Coryne, API ZYM, and API 50 CH strips were used as described by the manufacturer (bioMérieux). Conventional biochemical tests or API panels were incubated at 28 °C. Growth at 37 °C and 45 °C was assessed after incubation in ISP 2 medium [35] for 5 days. For analyses of chemotaxonomic characteristics, cells of Nocardia strains were grown in tryptic soy broth (TSB) for 3 days at 30 °C and harvested by centrifugation. Isoprenoid quinones were extracted by using the integrated procedure of Minnikin et al. [36] and analyzed using liquid chromatography mass spectrometry (LCMS; model LCMS-8030 and LC-20AD; Shimadzu) equipped with a Senshu-Pak Pegasil ODS-SP-100 column (100 × 2.0 mm i.d.; Senshu Scientific, Tokyo, Japan). Methanol–isopropanol was used as the mobile phase (34% isopropanol, 60 min) at the flow rate of 0.2 ml min–1 with ultraviolet detection at 275 nm. The preparation and analysis of cellular fatty acid methyl esters were performed using the protocol of the MIDI Sherlock Microbial Identification System [37] and a gas chromatograph (6890N; Agilent Technologies) with Sherlock MIDI software (version 6.2) and the TSBA6 database (version 6.2). Summed feature 3 detected in MIDI system was analyzed by GC/MS (6890N; Agilent Technologies).
Result and discussion
The genome sizes of Nocardia strains used in this study ranged from 6.00 (N. paucivorans) to 10.52 Mb (N. miyunensis), with an average of 7.86 Mb (Table 1). N. vinacea demonstrated the lowest DNA G+C content (65.5 mol%), whereas N. harenae demonstrated the highest (72.0 mol%).
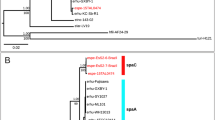
The similarities of concatenated atpD–groL1–groL2–recA–rpoA–secY–sodA–ychF nucleotide (nt) sequences (total 9680 nt) among the tested strains ranged from 83.90% (between N. flavorosea and N. inohanensis) to 99.65% (between N. cummidelens and N. soli). The NJ phylogenetic tree derived from the nt sequences concatenated the eight housekeeping genes (Fig. 1). Of the 71 branches in the phylogenetic tree, 41 were supported by 100% bootstrap value and 46 were supported by the NJ phylogenetic tree based on the amino acid sequences (aa) that concatenated the eight housekeeping genes. There was excellent correlation between the phylogenetic relationships observed between species in the individual clades and those observed in a phylogenetic study of 190 clinical, 36 type, and 11 reference strains based on five-locus MLSA [6]. Phylogenetically, N. cerradoensis, N. mikamii, N. kruczakiae, N. aobensis, N. nova, N. elegans, and N. africana form a coherent clade. This clade included the species reported as N. asteroides Type III Drug Susceptibility Pattern [4, 38]. Their MLSA similarities ranged from 97.67% to 99.13%. Moreover, N. gamkensis, N. exalbida, and N. arthritidis (98.13 to 98.96% MLSA (nt) similarities); N. cummidelens, N. soli, and N. salmonicida (98.61 to 99.65% MLSA (nt) similarities); N. coubleae and N. ignorata (99.29% MLSA (nt) similarities); and N. brasiliensis and N. vulneris (99.01% MLSA (nt) similarities) also formed coherent clades. N. coubleae and N. ignorata, and N. arthritidis, N. gamkensis and N. exalbida were previously reported by McTaggart et al. [6] as two sets of type strains that form distinct clusters. Each clade was sustained in the ML and MP phylogenetic trees.
Neighbor-joining phylogenetic tree of the genus Nocardia based on MLSA using concatenated atpD–groL1–groL2–recA–rpoA–secY–sodA–ychF gene sequences (9680 nucleotides (nt)). Numbers at nodes are bootstrap values based on 1000 resamplings (only values >70% are indicated). Asterisks indicate that the clades were recovered in the neighbor-joining tree using amino acid sequences, and daggers indicate that the clades were recovered in both the maximum-likelihood (nt) and the maximum-parsimony (nt) trees. Bar, 0.02% sequence divergence
The dDDH relatedness among N. cerradoensis, N. mikamii, N. kruczakiae, N. aobensis, N. nova, N. elegans, and N. africana ranged from 45.6% to 75.1%. N. nova and N. elegans showed 75.1% relatedness (by dDDH), which is higher than the 70 % cut-off point of DDH relatedness for the assignment of bacterial strains to the same genomic species [39]. This relatedness between N. nova and N. elegans was supported by laboratorial DDH (88 to 91%). Both G+C contents were 67.9 mol%. These strains assimilate glucose, but not arabinose, citrate, galactose, myo-inositol, mannitol, rhamnose, sorbitol, trehalose, or xylose. They however hydrolyze urea [40, 41]. The phenotypic characteristics determined in this study were similar for N. nova and N. elegans (Table 2). The similarity between the 16 S rRNA gene sequences of N. nova and N. elegans was only 98.2%, although the dDDH relatedness were 75.1%. Kim et al. [19] reported that 98.65% of 16S rRNA gene sequence similarity is the threshold for recognizing novel species using dDDH instead of laboratorial DDH, but the cut-off may not guarantee different genomic species status because there are exceptional cases owing to higher levels of intraspecies divergence of 16S rRNA gene sequences. Furthermore, the similarities between N. nova and N. elegans are within the threshold range (98.2% and 99.0% of 16S rRNA gene sequence similarity), which is on the boundary for species delineation [22]. The major cellular fatty acids were C16:0 (34.4–38.1%), C18:1 ω9c (21.0–22.4%), C18:0 10-methyl (tuberculostearic acid (TBSA)) (17.0–18.8%), and C18: 0 (13.5–15.9%). Detailed fatty acid components are presented in Table S1. The predominant menaquinone of N. nova and N. elegans and N. soli was MK-8 (H 4ω-cycl).
The genomic relationships among N. aobensis, N. cerradoensis, and N. kruczakiae ranged from 59.8% to 65.3% relatedness (by dDDH). This finding correlated with that of Kageyama et al. [42] who reported that the DDH relatedness between N. aobensis and N. cerradoensis was 53% to 59%.
N. exalbida and N. gamkensis formed a coherent clade in MLSA phylogenetic tree, and showed 73.7% relatedness by dDDH. Although the 16S rRNA gene sequences similarity was high (99.4%), they had not been compared with each other in their original papers because they were proposed around the same time [43, 44]. The relatedness was also supported by laboratorial DDH (76 to 89%). Their GC content fell within the narrow range of 68.4 to 68.6 mol%. N. gamkensis was reported to weakly utilize galactose and mannose, and to grow at 45 °C [44]. However, their utilization and growth at 45 °C were negative for N. gamkensis similar to N. exalbida [43] (Table 2). The major cellular fatty acids were C16:0 (30.0–34.6%), C18: 0 (16.9–23.9%), C18:1 ω9c (16.7–20.4%), and C18:0 10-methyl (TBSA) (12.6–15.7%). Detailed fatty acid components are presented in Table S1. The predominant menaquinone of N. exalbida and N. gamkensis was MK-8 (H 4ω-cycl).
N. arthritidis isolated from the clinical samples of Japanese patients [45] was shown to be related to N. exalbida/N. gamkensis (62.2% to 62.4% relatedness by dDDH), but can utilize ribose unlike N. exalbida and N. gamkensis.
N. cummidelens also showed 92.9% relatedness by dDDH with N. soli. N. salmonicida showed similarities ranging from 78.8% to 79.3% relatedness (by dDDH) with N. cummidelens and N. soli. It was confirmed by laboratorial DDH relatedness (75 to 90%). They had almost same G+C contents (67.0 to 67.1 mol%). N. cummidelens and N. soli were reported as novel species forming a monophyletic clade in the 16S rRNA gene sequence tree together with N. salmonicida [46]. N. cummidelens and N. soli had 100% similarity of the 16S rRNA gene sequence, and they showed 99.5% similarities with N. salmonicida. N. soli reportedly utilizes rhamnose [46], and N. salmonicida utilizes mannitol and sorbitol [47]. In this study, N. soli, N. cummidelens, and N. salmonicida could not utilize rhamnose, mannitol, and sorbitol. Other phenotypic characteristics were also similar among N. salmonicida, N. soli, and N. cummidelens (Table 2). The major cellular fatty acids were C16:0 (33.9–36.1%), C18:0 10-methyl (TBSA) (19.0–20.7%), C18:1 ω9c (13.8–21.5%), and C16:1 ω7c (9.7–15.4%). Detailed fatty acid components are presented in Table S1. The predominant menaquinone of N. salmonicida, N. cummidelens, and N. soli was MK-8 (H 4ω-cycl).
N. coubleae and N. ignorata showed 74.8% relatedness by dDDH. The laboratorial DDH between N. coubleae and N. ignorata tested in this study was also 79%, although this has previously been reported as 26 % [48]. The 16S rRNA gene sequence similarity was 99.4%. Their GC content fell within the narrow range of 67.7 to 67.9 mol%. N. ignorata was reported to grow at 45 °C [49]. However, it could not grow at this characteristic temperature in this study. A similar phenomenon was observed with vcoubleae (Table 2). Although N. ignorata was reported possessing MK-8(H6) or MK-8(H4cycl) as major menaquinone, it was not MK-8(H6) but MK-8(H4cycl) as with that of N. coubleae in this study. The major cellular fatty acids were C16:0 (27.9–38.5%), C18:0 10-methyl (TBSA) (11.8–11.9%), C18: 0 (2.6–16.8%), C16:1 ω7c (28.4–30.6%), and C18:1 ω9c (10.5–14.2%). Detailed fatty acid components are presented in Table S1.
N. brasiliensis and N. vulneris showed 65.7% relatedness (by dDDH). This was consistent with the result of laboratorial DDH by Lasker et al. [50]. They reported that N. vulneris was readily distinguished phenotypically from N. brasiliensis, although it was in a transitional gray zone near the 70% threshold of DDH [50].
In conclusion, on the basis of genotypic and phenotypic data, it is evident that N. soli and N. cummidelens should be reclassified as later heterotypic synonyms of N. salmonicida, N. gamkensis as a later heterotypic synonym of N. exalbida, N. coubleae as a later heterotypic synonym of N. ignorata, and N. elegans as a later heterotypic synonym of N. nova.
Emended description of Nocardia salmonicida (ex Rucker 1949) Isik et al. 1999
The description is as that of Isik et al. [47] with the following amendments. Growth may occur at 37 °C. Positive for (in API ZYM and API coryne) catalase, urea hydrolysis, alkaline phosphatase, esterase (C-4), esterase lipase (C-8), α-glucosidase, and β-glucosidase. Utilizes d-glucose, glycerol, d-fructose and N-acetyl-glucosamine. MK-8(H4cycl) is the predominant menaquinone. The major fatty acids are C16:0, C18:0 10-methyl, C18:1 ω9c, and C16:1 ω7c.
The type strain, NBRC 13393T (=ATCC 27463T=CBS 694.72T=CIP 104517T=DSM 40472T=JCM 4826T=NRRL B-2778T=NRRL B-12385T), was a fish pathogen isolated from blueblack salmon (Oncorhynchus nerka) [47]. The names Nocardia soli (NBRC 100376T) and Nocardia cummidelens (NBRC 100378T) are later heterotypic synonyms.
Emended description of Nocardia nova Tsukamura 1983
The description is as that of Tsukamura [40] with the following amendments. Positive for (in API ZYM and API coryne) catalase, nitrate reduction, alkaline phosphatase, acid phosphatase, esculin hydrolysis,α-glucosidase, β-glucosidase, and phosphohydrolase. MK-8(H4cycl) is the predominant menaquinone. The major fatty acids are C16:0, C18:1 ω9c, C18:0 10-methyl (TBSA), and C18:0.
The type strain, NBRC 15556T (=Tsukamura 23095T=R.E. Gordon R443T=ATCC 33726T=CCUG 45939T=CIP 104777T=DSM 44481T=JCM 6044T=VKM Ac-1971T), was a lung pathogenic bacterium [40]. The name Nocardia elegans (NBRC 108235T) is a later heterotypic synonym.
Emended description of Nocardia exalbida Iida et al. 2006
The description is as that of Iida et al. [43]. with the following amendments. Growth may occur at 37 °C. Positive for (in API ZYM and API coryne) catalase, nitrate reduction, alkaline phosphatase, acid phosphatase, esterase (C-4), and esterase lipase (C-8). Utilizes d-glucose, glycerol, d-fructose, and N-acetyl-glucosamine. MK-8(H4cycl) is the predominant menaquinone. The major fatty acids are C16:0, C18: 0, C18:1 ω9c, and C18:0 10-methyl (TBSA).
The type strain, NBRC 100660T (=DSM 44883T=IFM 0803T=JCM 12667T), was isolated from the bronchoalveolar lavage of an immunocompromised patient with lung abscess, in Chiba, Japan [43]. The name Nocardia gamkensis (NBRC 108242T) is a later heterotypic synonym.
Emended description of Nocardia ignorata Yassin et al. 2001
The description is as that of Yassin et al. [49] with the following amendments. The predominant menaquinones is MK-8(H4cycl). Positive for (in API ZYM and API coryne) catalase, nitrate reduction, urea hydrolysis, gelatin hydrolysis, esterase (C-4), esterase lipase (C-8), and α-glucosidase. Utilizes d-glucose and N-acetyl-glucosamine. MK-8(H4cycl) is the predominant menaquinone. The major fatty acids are C16:0, C18:0 10-methyl (TBSA), C18: 0, C16:1 ω7c, and C18:1 ω9c.
The type strain, NBRC 108230T (=CCUG 48296T=DSM 44496T=IMMIB R-1434T=JCM 11764T=NRRL B-24141T), was originally identified as Mycobacterium sp. from a specimen sent to the clinical microbiological laboratory [49]. The name Nocardia coubleae (NBRC 108252T) is a later heterotypic synonym.
References
Trevisan, V. I. generi e le specie delle Batteriacee. Zanaboni and Gabuzzi, Milano, 1889, 1-35.
Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–91.
Goodfellow M, Maldonado LA, Genus I. Nocardia Trevisan 1889AL. In: Goodfellow M et al. (eds). Bergey’s manual of systematic bacteriology, 2nd edn, Vol. 5, the Actinobacteria, part A. Springer: New York, 2012, pp 376–419.
Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev. 2006;19:259–82.
Eshraghi SS. Molecular typing of Nocardia species. J Med Bacteriol. 2012;1:38–45.
McTaggart LR, Richardson SE, Witkowska M, Zhang SX. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J Clin Microbiol. 2010;48:4525–33.
Wang X, et al. Reverse line blot hybridization and DNA sequencing studies of the 16S-23S rRNA gene intergenic spacer regions of five emerging pathogenic Nocardia species. J Med Microbiol. 2010;59:548–55. Pt 5
Chun J, Rainey FA. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol. 2014;64(Pt 2):316–24.
Sangal V, Nieminen L, Tucker NP, Hoskisson PA. Revolutionising systematics through next-generation sequencing. In: Goodfellow M, Sutcliffe IC, Chun J (eds). Methods in microbiology: bacterial taxonomy, Vol. 41. Elsevier: London, 2014, pp 75–101.
Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52(Pt 3):1043–7.
Rong X, Huang Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol. 2012;35:7–18.
Wink J, et al. ‘Streptomyces caelicus’, an antibiotic-producing species of the genus Streptomyces, and Streptomyces canchipurensis Li et al. 2015 are later heterotypic synonyms of Streptomyces muensis Ningthoujam et al. 2014. Int J Syst Evol Microbiol. 2017;67:548–56.
McDowell A, Barnard E, Liu J, Li H, Patrick S. Proposal to reclassify Propionibacterium acnes type I as Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes type II as Propionibacterium acnes subsp. defendens subsp. nov. Int J Syst Evol Microbiol. 2016;66:5358–65.
Fidalgo C, Riesco R, Henriques I, Trujillo ME, Alves A. Microbacterium diaminobutyricum sp. nov., isolated from Halimione portulacoides, which contains diaminobutyric acid in its cell wall, and emended description of the genus Microbacterium. Int J Syst Evol Microbiol. 2016;66:4492–4500.
Komaki H, et al. Genome analysis-based reclassification of Streptomyces endus and Streptomyces sporocinereus as later heterotypic synonyms of Streptomyces hygroscopicus subsp. hygroscopicus. Int J Syst Evol Microbiol. 2017;67:343–5.
Tang B, et al. A systematic study of the whole genome sequence of Amycolatopsis methanolicastrain 239(T) provides an insight into its physiological and taxonomic properties which correlate with its position in the genus. Synt Syst Biotechnol. 2016;1:169–86.
Borriss R, Rueckert C, Blom J, Bezuidt O, Reva O, Klenk H-P. Whole genome sequence comparisons in taxonomy. Methods Microbiol. 2011;38:409–36.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. Pt 1
Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51. Pt 2
Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–72.
Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–9.
Meier-Kolthoff JP, Goker M, Sproer C, Klenk HP. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–8.
Meier-Kolthoff JP, Klenk HP, Goker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–6. Pt 2
Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–31.
Meng X, et al. Actinomyces liubingyangii sp. nov. isolated from the vulture Gypaetus barbatus. Int J Syst Evol Microbiol. 2017;67:1873–9.
Mohammadipanah F, Montero-Calasanz MD, Schumann P, Sproer C, Rohde M, Klenk HP. Promicromonospora kermanensis sp. nov., an actinobacterium isolated from soil. Int J Syst Evol Microbiol. 2017;67:262–7.
Rottig A, et al. Streptomyces jeddahensis sp. nov., an oleaginous bacterium isolated from desert soil. Int J Syst Evol Microbiol. 2017;67:1676–82.
Sangal V, et al. Next-generation systematics: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep. 2016;6:38392.
Tamura T, et al. A genome sequence-based approach to taxonomy of the genus Nocardia. Antonie Van Leeuwenhoek. 2012;102:481–91.
Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinform. 2008;9:376.
Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol. 2012;194:6970–1.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9.
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60.
Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol. 1989;39:224–9.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Minnikin DE, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–41.
Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. Newark: Microbial ID, Inc.; 1990.
Wallace RJ Jr., Steele LC, Sumter G, Smith JM. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob Agents Chemother. 1988;32:1776–9.
Wayne LG, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4.
Tsukamura M. Numerical analysis of the taxonomy of Nocardiae and Rhodococci. Microbiol Immunol. 1982;26:1101–19.
Yassin AF, Brenner S. Nocardia elegans sp. nov., a member of the Nocardia vaccinii clade isolated from sputum. Int J Syst Evol Microbiol. 2005;55(Pt 4):1505–9.
Kageyama A, Suzuki S, Yazawa K, Nishimura K, Kroppenstedt RM, Mikami Y. Nocardia aobensis sp. nov., isolated from patients in Japan. Microbiol Immunol. 2004;48:817–22.
Iida S, et al. Nocardia exalbida sp. nov., isolated from Japanese patients with nocardiosis. Int J Syst Evol Microbiol. 2006;56(Pt 6):1193–6.
le Roes M, Meyers PR. Nocardia gamkensis sp. nov. Antonie Van Leeuwenhoek. 2006;90:291–8.
Kageyama A, et al. Nocardia arthritidis sp. nov., a new pathogen isolated from a patient with rheumatoid arthritis in Japan. J Clin Microbiol. 2004;42:2366–71.
Maldonado L, Hookey JV, Ward AC, Goodfellow M. The Nocardia salmonicida clade, including descriptions of Nocardia cummidelens sp. nov., Nocardia fluminea sp. nov. and Nocardia soli sp. nov. Antonie Van Leeuwenhoek. 2000;78:367–77.
Isik K, Chun J, Hah YC, Goodfellow M. Nocardia salmonicida nom. rev., a fish pathogen. Int J Syst Bacteriol. 1999;49(Pt 2):833–7.
Rodriguez-Nava V, Khan ZU, Potter G, Kroppenstedt RM, Boiron P, Laurent F. Nocardia coubleae sp. nov., isolated from oil-contaminated Kuwaiti soil. Int J Syst Evol Microbiol. 2007;57(Pt 7):1482–6.
Yassin AF, Rainey FA, Steiner U. Nocardia ignorata sp. nov. Int J Syst Evol Microbiol. 2001;51(Pt 6):2127–31.
Lasker BA, et al. Nocardia vulneris sp. nov., isolated from wounds of human patients in North America. Antonie Van Leeuwenhoek. 2014;106:543–53.
Acknowledgements
This study was partly supported by the commissioned project from the Japan Patent Office.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that this research was conducted without any financial and commercial relationships with profit-making corporations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tamura, T., Ohji, S., Ichikawa, N. et al. Reclassification of Nocardia species based on whole genome sequence and associated phenotypic data. J Antibiot 71, 633–641 (2018). https://doi.org/10.1038/s41429-018-0043-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-018-0043-1
This article is cited by
-
Genome insights into the pharmaceutical and plant growth promoting features of the novel species Nocardia alni sp. nov
BMC Genomics (2022)
-
Assessment of VITEK® MS IVD database V3.0 for identification of Nocardia spp. using two culture media and comparing direct smear and protein extraction procedures
European Journal of Clinical Microbiology & Infectious Diseases (2020)
-
Bioactive molecules fromNocardia: diversity, bioactivities and biosynthesis
Journal of Industrial Microbiology and Biotechnology (2019)