Abstract
4,4′-bond secalonic acid D (4,4′-SAD) is a known compound isolated from the marine-derived fungus Penicillium oxalicum. No study about the antitumor effect of this compound has been reported, except for a few focusing on its bactericidal properties. Herein, we performed an in vitro biology test and found that 4,4′-SAD stimulated the apoptosis of tumor cells in the human hepatocellular carcinoma cell lines PLC/PRF/5 and HuH-7 by activating caspase-3, caspase-8, caspase-9, PARP, p53, and cyclin B1, as well as by regulating the Bax/Bcl-2 ratio. In vivo studies showed that 4,4′-SAD had antitumor efficacy in H22 cell xenograft model. Immunohistochemical analysis revealed that 4,4′-SAD could regulate Bax expression, which is a biomarker of tumor growth. In summary, 4,4′-SAD significantly inhibited tumor growth both in vivo and in vitro.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is known as the sixth most widespread cancer and the third primary reason of cancer-related deaths in the world [1]. Chronic infection with hepatitis B and C, fatty liver disease, aflatoxin, and alcohol abuse are the principal factors leading to HCC [2]. HCC is characterized by concealed onset, rapid progress, poor prognosis, and ascertained advanced stage upon first diagnosis [3]. Thus far, the clinically effective therapies for HCC include surgery, transarterial chemoembolization, radiotherapy, immunotherapy, chemotherapy, molecular-targeted therapy, and other systemic treatments [4]. However, most of the systemic chemotherapeutics have no efficacy in the treatment of HCC, leading to an urgent need to identify novel therapy drugs [5].
Nature-derived anticancer drugs, such as taxane, bleomycin, and vinca alkaloids, have played a significant role in the history of cancer therapy. However, after 50 years of intensive screening from plants and terrestrial microbes, the pace of natural product discovery and development has gradually declined over the last two decades. Fortunately, since the early 1990s, the number of marine source-purified compounds that have been shown to affect human cancer has increased dramatically [6]. In the process of screening bioactive secondary metabolites secreted from marine fungus, we isolated a strain named Penicillium oxalicum from sediments in the Langqi island of Fujian coast. After a series of fermentation, extraction, and purification steps, 4,4′-SAD was obtained through HPLC and determined to have the same structure as reported by spectroscopic methods (NMR and MS) [7]. Several studies have reported the remarkable antitumor activity of its analog SAD (2,2′-bond) through different mechanisms in several cell lines, including embryonic palatal mesenchymal cell [8], leukemia cell [9], ABCB1-, ABCC1- and ABCG2-overexpressing multidrug resistance cells [10], GH3 cells [11], and MCF-7 cells [12]. However, no literature about the antitumor effect of 4,4′-SAD has been published yet, except for a few bactericidal reports [7]. Our studies proved that 4,4′-SAD could inhibit HCC cell proliferation and induce apoptosis through caspase-dependent pathway in vitro and in vivo, indicating the potential value of 4,4′-SAD in HCC treatment.
Materials and methods
Fungal materials
The fungus P. oxalicum was purified from marine sediments collected from the southeast coastal region of China. P. oxalicum was identified through ITS (internal transcribed spacer) analysis by Sunbiotech Co. Ltd. The voucher specimen was preserved in the China Center for Type Culture Collection (no. M2013714). The producing strain was prepared on Martin medium and stored at 4 °C.
Fermentation and extraction
The fungus was cultured under static conditions as we described earlier [13]. In a typical procedure, the liquid medium included NaCl (1.5%), maltose (2%), glucose (1%), mannitol (2%), monosodium glutamate (1%), MgSO4•7H2O (0.03%), KH2PO4 (0.05%), and yeast extract (0.3%). After being cultured at 28 °C for 30 days (60 L), the broth was filtered to separate supernatant from mycelia. The mycelia were extracted with acetone three times and concentrated to produce an aqueous solution. The aqueous solution was extracted with EtOAc three times and concentrated to produce an extract (36.5 g).
Purification
The mycelia extract was passed through a Si gel column to yield five fractions (A–E). Fraction C (12.7 g) was separated on a Sephadex column (LH-20; CH2Cl2/MeOH = 1:2) to yield four subfractions (C1–C4). Subfraction C3 (4.9 g) was finally purified by HPLC (55% CH3CN and 0.1% TFA), yielding 4,4′-SAD (1.1 g).
4,4′-SAD: yellow powder (CHCl3); [α]20D –123.3° (c 0.3, pyridine); 1H and 13C-NMR (Table S1); HRESIMS (m/z: 661.1531 [M + Na]+, calcd. for C32H30NaO14, 661.1533); HRESIMS (m/z: 637.1573 [M–H]–, calcd. for C32H29O14, 637.1563).
Culture of cell lines
All the human cancer cell lines were purchased from Shanghai Cell Resource Center (Shanghai, China). Murine tumor cell lines, including liver ascites tumor H22, were obtained from the Cancer Metastasis Alert and Prevention Center, Fuzhou University. All cell lines were cultured in RPMI-1640, EMEM, or DMEM (Hyclone, USA) supplemented with 10% fetal bovine serum (Hyclone, USA) at 37 °C in 5% (v/v) CO2 humidified atmosphere.
Animals
Six-week-old Kunming mice (18–20 g) were purchased from Minhou County Wu Experimental Animal Trade Co., Ltd. (Fuzhou, China). All animals used in the investigation were handled in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996) and approved by the Animal Care and Use Committee of Institute of Biomedical and Pharmaceutical Technology, Fuzhou University. All mice were killed with excess amounts of anesthetic.
Cell viability analysis
Cell viability was assessed by the WST-1 cell proliferation and cytotoxicity assay kit (Roche, Indianapolis, IN, USA) as previously described [14, 15]. In a typical procedure, cells (4 × 103 per well) were plated in 100 μL culture medium per well in 96-well plates for 24 h and then treated with gradient concentrations of 4,4′-SAD or SAD for 72 h. After incubated with DMEM containing 10% WST-1 for another 4 h, the cells were tested with an ELISA reader (TECAN Infinite M200 Pro, Switzerland) at 450 nm. Each concentration had five repetitive wells, and each experiment was repeated at least three times. The IC50 values were calculated using Graphpad Prism software.
AO/EB staining
Cells (2 × 105 per well) were cultured with different concentrations of 4,4′-SAD for 48 h, and then stained with a 1:1 mixed solution of (100 μg/mL) AO (acridine orange) and (100 μg/mL) EB (ethidium bromide) for 2 min in darkness. Cell morphology was observed under a fluorescent microscope (Ti-E, Nikon, Tokyo, Japan).
Cell cycle assay
Cell cycle assay was performed by flow cytometry as previously described [16]. In a typical procedure, cells were collected, fixed, and permeabilized with cold 70% (v/v) ethanol at 4 °C overnight. After staining with PI (Propidium Iodide) at 37 °C, cells were determined by flow cytometry (Beckman Coulter FC500, Brea, CA, USA), and the image was analyzed by Flowjo software.
Annexin V/PI staining
Apoptosis and necrosis cells were detected with the Annexin V FLUOS staining kit (Roche, Mannheim, Germany) as previously described [16]. In a typical procedure, cells were collected, washed, and incubated with Annexin V-EGFP buffer for 15 min. Furthermore, cells were resuspended in 200 μL mixed solution with PI and tested by flow cytometry (Beckman Coulter FC500, Brea, CA, USA). The pan-caspase inhibitor, Z-VAD-FMK (Beyotime, Haimen, China), was added to cells at the final concentration of 50 μM before 4,4′-SAD treatment. Data were analyzed by Flowjo software.
Real-time cell analysis proliferation assay
The DP version of the xCELLigence real-time cell analyzer RTCA (ACEA, San Diego, CA, USA) was used to monitor real-time cell proliferation. The specific experiment methods were performed as previously described [16].
Western blot assay
Cells were collected and lysed with RIPA buffer. The supernatant was separated with 12% SDS-PAGE through electrophoresis (Bio-Rad, Richmond, CA, USA). The protein sample was transferred to the NC membrane, which was blocked with 5% (w/v) milk-PBST for 1 h and incubated with specific primary antibodies (including anti-Bax, Bcl-2, caspase-3, caspase-8, caspase-9, PARP, cyclin B1, p53, and β-actin antibodies; all are anti-rabbit and 1:1000 dilution (Cell Signaling Technology, USA) in 5% (w/v) milk-PBST overnight at 4 °C). The membranes were subsequently washed three times every 10 min in PBST and incubated with peroxidase-conjugated anti-rabbit IgG (1:5000, BOSTER, China) secondary antibody. Protein bands were detected by FluorChem E digital darkroom system (Protein Simple, Santa Clara, USA).
RNA extraction and quantitative real-time PCR analysis
Total RNA was isolated from cells with TRIzol reagent (Life Technologies, Carlsbad, CA, USA), and messenger RNA (mRNA) was synthesized to complementary DNA with the Goscript™ Reverse Transcriptase System (Promega, WI, USA). Quantitative PCR was detected using Light Cycler 480 II (Roche, Switzerland) with the miScript SYBR Green PCR kit (QIAGEN, Hilden, Germany). The oligonucleotide primer pairs used in the present study are listed in Table S2.
In vivo H22 s.c. tumor model
To investigate the effects of 4,4′-SAD on tumor growth in vivo, we established a xenograft tumor model [17, 18]. In a typical procedure, each mouse was injected with 1 × 106 H22 hepatocarcinoma cells s.c. into the right flank. Mice were randomly divided into four groups (n = 6 for each group). Dry compounds were suspended in 1% carboxy methyl cellulose–Na (CMC–Na, purchased from Sinoharm Chemical Reagent Co., Ltd., China) for immediate use. Mice were treated with 200 μL 1% CMC–Na (negative control) and 4,4′-SAD (10, 20, and 40 mg/kg) by gavage every day from days 2 to 14. The weight and tumor volumes were recorded with electronic scales and slide caliper, respectively, every other day until mice were killed on day 14. The massive tumor was peeled off, and pictures were taken. Tumors and normal tissues such as heart, liver, spleen, kidney, and lung were extracted. After washing with physiological saline, they were fixed in formalin and prepared in sections for histological observation. Tumor inhibition rate (%) = (mean tumor weight of control group − mean tumor weight of treated group)/mean tumor weight of control group × 100%.
Immunohistochemistry
Tumor tissues in Kunming mice were collected, fixed, paraffin embedded, cut into pieces, and incubated with anti-Bax antibody [19]. The expression of Bax was determined with the DAB assay kit (ZSGB-Bio, Beijing, China). All sections were photographed using a ×400 normal light microscope.
Statistical analysis
All results are expressed as mean ± SD of triplicate samples. P values <0.05 were considered significantly different. All statistical analyses were conducted using SPSS 13.0 software.
Results
Purification and structural elucidation
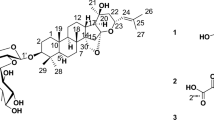
The mycelia extract of P. oxalicum was consecutively separated on silica gel, sephadex LH-20, and preparative HPLC columns to obtain the bioactive compound having the same molecular formula of C32H30O14 with known SAD by positive and negative HRESIMS. However, different retention times on HPLC, 1H-NMR, and 13C-NMR suggested that this compound had different structures from known SAD. The compound’s structure was accurately confirmed by a detailed elucidation through COSY, HMBC, and NOE correlations of NMR (Figure S1), as well as by comparing its specific rotatory power and 1H and 13C-NMR data (Table S1) with literature values [7]. The compound was finally identified as 4,4′-SAD (Fig. 1).
4,4′-SAD suppressed the proliferation of multiple cancer cell lines
To compare the cytotoxic activity of 4,4′-SAD with SAD, we initially investigated the proliferation inhibition effect against 22 human cancer cell lines and the murine hepatocarcinoma cell line H22 after the treatment with gradient concentrations of 4,4′-SAD or SAD for 72 h. As shown in Figure S2, both 4,4′-SAD and SAD significantly inhibited cell proliferation in all cell lines, but 4,4′-SAD had stronger cytotoxic effects than SAD. The average IC50 values of 4,4′-SAD and SAD to 22 human cell lines were 1.026 and 1.353 µM, respectively (Table 1). Similarly, the IC50 of 4,4′-SAD and SAD to H22 cells were 1.211 and 1.007 µM (Table 1). Considering the high fatality rate of HCC, we finally selected human cancer cell lines PLC/PRF/5 and HuH-7 for further mechanism study in vitro and murine cancer cell line H22 for subsequent study in vivo.
We adopted an real-time cell analysis (RTCA) assay to accurately characterize the anti-proliferative potential of 4,4′-SAD in PLC/PRF/5 and HuH-7 cells. As shown in Fig. 2a, b, the cell proliferation under the gradient concentrations of 4,4′-SAD treatment was recorded continuously for about 100 h through xCELLigence RTCA system. Results clearly confirmed that 4,4′-SAD could inhibit cell growth from the tendency of real-time cell growth curves.
4,4′-SAD-induced apoptosis in liver cancer cells
First, we adopted AO/EB staining to explore whether the 4,4′-SAD could induce cancer cell apoptosis or not. As shown in Fig. 3a, higher concentration of 4,4′-SAD could lead to a sharp increase of apoptotic cells. The apoptotic rates were 16.7%, 36.4%, and 46.6% in PLC/PRF/5 and 10.8%, 21.6%, and 35.8% in HuH-7 cell line when treated with 1, 2, and 4 μM 4,4′-SAD, respectively. Second, the cell cycle was analyzed to reveal the possible mechanism of cell growth inhibition. As shown in Fig. 3b, when treated with gradient concentrations of 4,4′-SAD (0, 1, 2, and 4 μM), the percentage of sub G1 (apoptosis peak) increased from 9.3% to 21.3%, 31.8%, and 39.0% in PLC/PRF/5 and from 5.1% to 17.0%, 33.5%, and 35.0% in HuH-7 cell line, respectively. Results of Annexin V/PI double staining (Fig. 3c) further revealed that the apoptotic rates increased from 17.8 and 39.8% to 42.8% in PLC/PRF/5 and from 10.4 and 18.8% to 27.3% in HuH-7 cell line when treated with 1, 2, and 4 μM of 4,4′-SAD. Taken together, these results indicated that 4,4′-SAD could induce clear apoptosis in liver cancer cell lines.
4,4′-SAD-induced significant apoptosis in PLC/PRF/5 and HuH-7 cells. (a) AO/EB staining of PLC/PRF/5 and HuH-7 cells treated with 4,4′-SAD (0, 1, 2, and 4 µM) for 48 h; (b) cell cycle test of PLC/PRF/5 and HuH-7 cells treated with 4,4′-SAD (0, 1, 2, and 4 µM) for 48 h by flow cytometry; (c) Annexin V/PI test of PLC/PRF/5 and HuH-7 cells treated with 4,4′-SAD (0, 1, 2, and 4 µM) for 48 h
To elucidate the mechanism of 4,4′-SAD-induced apoptosis, the apoptosis-related proteins and genes were detected by western blot and real-time PCR. The caspase family plays an important role in the process of apoptosis and is involved in almost all signal transduction pathways during apoptosis. Caspases-8 and -9 are early-stage initiators of cell apoptosis, whereas caspase-3 is a late-stage executor [20]. Moreover, PARP induced by DNA damage repair can be cut by caspase-3 and lose activity. As shown in Fig. 4a, b, d, e, PARP, caspase-3, caspase-8, and caspase-9 were cleaved and activated in both PLC/PRF/5 and HuH-7 cells after exposure to the indicated concentrations of 4,4′-SAD for 48 h. To further confirm whether 4,4′-SAD-induced apoptosis was dependent on caspase, we treated cells with or without 50 μM of the pan-caspase inhibitor Z-VAD-FMK together with 4 μM of 4,4′-SAD for 48 h. As the flow data showed, Z-VAD-FMK decreased 4,4′-SAD-induced apoptotic rate from 57.2 to 13.5% in PLC/PRF/5 cells and from 23.7 to 15.2% in HuH-7 cells, suggesting that Z-VAD-FMK inhibited 4,4′-SAD-induced apoptosis in both PLC/PRF/5 and HuH-7 cells (Fig. 5).
Expression of apoptosis-related proteins and genes in PLC/PRF/5 and HuH-7 cells treated with 4,4′-SAD. (a, d) Western blot of apoptosis-related proteins in PLC/PRF/5 and HuH-7 cells treated with 0, 1, 2, and 4 µM 4,4′-SAD for 48 h; (b, e) levels of apoptosis-related proteins analyzed by densitometry and represented as percentage compared with the control; (c, f) levels of apoptosis-related genes in PLC/PRF/5 and HuH-7 cells treated with 0, 1, 2, and 4 µM 4,4′-SAD for 48 h analyzed by RT-qPCR (loading control: GAPDH)
Z-VAD-FMK inhibited 4,4′-SAD-induced apoptosis. (a) PLC/PRF/5 and HuH-7 cells were incubated with 4 μM of 4,4′-SAD in the presence or absence of 50 μM of the caspase inhibitor, Z-VAD-FMK for 48 h, and then analyzed with Annexin V/PI staining by flow cytometry. (b) Densitometry of cell counts. Data are expressed as the mean ± SD (n = 3)
The Bcl-2/Bax ratio, which is the molecular switch mediating the mitochondrial apoptosis pathway, also decreased in a dose-dependent manner. At the same time, p53 and cyclin B1 proteins decreased with increasing 4,4′-SAD concentration. The mutations of p53 protein are known to normally result in the failure of interaction with DNA and the inactivation of normal p53 proteins expressed in cells, thereby preventing the transcriptional activation of genes involved in cell cycle arrest and apoptosis [14, 15, 21, 22]. Real-time PCR results further showed that alterations in these apoptosis-related genes at the mRNA expression level were almost consistent with those at the protein level after the treatment with 4,4′-SAD (Fig. 4c, f). These findings suggested that 4,4′-SAD may induce the apoptosis of PLC/PRF/5 and HuH-7 cells through multiple signaling pathways, including the activation of the caspase cascade signaling pathway, the regulation of Bax/Bcl-2 ratio, and the inhibition of aberrant protein expression in cancer cells.
4,4′-SAD inhibited tumor growth in vivo
To determine whether 4,4′-SAD could inhibit tumor growth in vivo, we established a xenograft tumor model using Kunming mice [17, 18]. We injected 1 × 106 H22 hepatocarcinoma cells s.c. into the right flank of 6-week-old mice and studied the antitumor efficacy of 4,4′-SAD by oral administration (Fig. 6a). As shown in Fig. 6b, tumor bulks in the 4,4′-SAD-treated group were significantly smaller than those in the control group, and the tumor inhibition rates of 4,4′-SAD treatment (10, 20, and 40 mg/kg) were 18.2%, 57.4%, and 72.6% compared with that of the control group, respectively. This finding suggested that 4,4′-SAD significantly inhibited tumor growth in dose- and time-dependent manners (Fig. 6b–e). Although the weight of mice slightly decreased with gradually increasing 4,4′-SAD concentration, no significant difference was found between the experiment and control groups (Fig. 6f). In addition, H&E staining showed that normal tissues such as heart, liver, spleen, lung, and kidney had no significant difference between the experiment and control groups (Fig. 6g), indicating the low toxicity of 4,4′-SAD. We also found that Bax increased in tumor tissues of 4,4′-SAD-treated mice through immunohistochemical staining (Fig. 6g), which may partly explain why 4,4′-SAD inhibited tumor growth in vivo. In general, these data revealed that 4,4′-SAD may exert good antitumor efficacy and safety in vivo.
4,4′-SAD inhibited tumor growths in vivo. (a) Schematic of the experimental protocol described in Materials and methods; (b, c) photographs and tumor weight of all tumors from Kunming mice treated by gradient concentration of 4,4′-SAD; (d, f) tumor volume and mice weight of each group measured every 2 days; (e) inhibition rate was quantitatively analyzed on day 14; (g) H&E staining of tumor and main viscera tissues of Kunming mice and Bax expression in tumor tissue by IHC
Discussion
To date, nine homologs of secalonic acids (secalonic acids A–I) have been isolated from natural source, and some of them were recently completely synthesized [13, 23, 24]. Some studies have shown that SAD, a teratogenic mycotoxin [25, 26], has teratogenicity to the offspring of pregnant mice, such as inducing cleft palate by inhibiting palatal shelf growth [27, 28], delaying olfactory discrimination and hindlimb grip behavior in males and females [29], and acting on the dose response of neurotoxic effects [30]. However, our data showed that 4,4′-SAD had no distinct side effects when used as an antitumor agent in vitro and in vivo. First, we accessed the cytotoxicity of 4,4′-SAD to normal cells, such as human foreskin fibroblast (HFF) cell, by WST-1 assay in vitro. In addition, the IC50 value of HFF was 26.6 μM (Table 1), which was almost 25 times that of PLC/PRF/5 and HuH-7 cells. Second, 4,4′-SAD-treated Kunming mice did not show significant toxic effects, such as a clear loss of body weight and even mortality at experimental doses. The data suggested that 4,4′-SAD preferred to target cancer cells rather than normal tissues, but the detailed mechanism requires further study.
Several anticancer agents exert their function by inducing apoptosis in tumor cells [31]. Apoptosis can be triggered by diverse cellular signals, such as intracellular signals formed in response to cellular stresses or triggered by DNA damage, or extracellular signals to cell surface receptors that activate the death-inducing signaling complex [32,33,34]. In general, we classified apoptosis into two major categories, and both the receptors (extrinsic) and mitochondrial (intrinsic) apoptotic pathways involved the activation of caspase-3 [35]. The activation of caspase-3, caspase-8, and caspase-9 showed in Fig. 4a, b strongly confirmed the occurrence of apoptosis. Notably, mitochondria play a unique role in apoptosis, whereas Bcl-2 family proteins are critical adjusters in mitochondria-mediated cell apoptosis [36,37,38]. Our data clearly showed that 4,4′-SAD treatment resulted in decreased Bcl-2/Bax protein ratio, which further demonstrated that 4,4′-SAD could promote apoptosis through the mitochondrial apoptotic pathway. In addition, DNA double-strand breaks and genome incompleteness generate a significant threat to cancer [39], but PARP can recruit the chromatin remodeler CHD2 to DNA damage and trigger the process of DNA damage repair [40]. The decrease of PARP and the increase of cleaved PARP after 4,4′-SAD treatment caused PARP to lose the function of repairing DNA damage and further accelerated cell apoptosis.
We also noticed that 4,4′-SAD downregulated p53 expression at both protein and mRNA levels (Fig. 4). Accumulating evidence supports that PLC/PRF/5 and HuH-7 cells express mutant p53 protein [41, 42] and neomorphic mutant p53 can promote apoptosis. Evidence from in vitro models, animals, and clinical studies also confirm that the expression of p53 point mutants is a crucial event in tumorigenesis that tips the balance toward overt malignant progression [43,44,45]. We proposed that the remarkable efficacy of 4,4′-SAD in inhibiting tumor may be partly due to the downregulation of p53 expression.
At last, it’s worth mentioning that 4,4′-SAD was found to be stable in dry powder (Figure S14), however unstable in solvent (Figures S12 and S13), which was also reported to exist in other secalonic acids, such as secalonic acid A (SAA) [46]. Although 4,4′-SAD took a longer time to reach the equilibrium than 4,4′-SAA, the final ratios of 2,2′-, 2,4′-, and 4,4′-linked variants between these secalonic acids were similar. It was interesting to find out that 4,4′-SAA showed average four times stronger bioactivity than that of SAA as reported [46], however 4,4′-SAD only showed a little stronger than SAD. Furthermore, in order to ensure the purity of 4,4′-SAD during our each experiment, this compound used for all the biological studies was sub-packaged into a lot of Eppendorf tubes and stored at −20 °C in dry powder. Each tube taken out of refrigerator was dissolved and used immediately. After each experiment, the residue of 4,4′-SAD was discarded and refused to reuse.
In conclusion, 4,4′-SAD as a compound with similar structure of SAD showed great antitumor activity by apoptosis induction with low toxicity and high safety. Thus, 4,4′-SAD can be a promising antitumor lead compound waiting for further exploration.
References
Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15:5–13.
Fitzmaurice C, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–27.
Amin A, Hamza AA, Bajbouj K, Ashraf SS, Daoud S. Saffron: a potential candidate for a novel anticancer drug against hepatocellular carcinoma. Hepatology. 2011;54:857–67.
Kubo S, Takemura S, Sakata C, Urata Y, Uenishi T. Adjuvant therapy after curative resection for hepatocellular carcinoma associated with hepatitis virus. Liver Cancer. 2013;2:40–6.
Ting CT, Li WC, Chen CY, Tsai TH. Preventive and therapeutic role of traditional chinese herbal medicine in hepatocellular carcinoma. J Chin Med Assoc. 2015;78:139–44.
Lordan S, Ross RP, Stanton C. Marine bioactives as functional food ingredients: potential to reduce the incidence of chronic diseases. Mar Drugs. 2011;9:1056–1100.
Kurobane I, Ving LC, Mcinnes AG. Secalonic acids and their use. German Pat. DE 3002761. 1980.
Dhulipala VC, Maddali KK, Welshons WV, Reddy CS. Secalonic acid D blocks embryonic palatal mesenchymal cell-cycle by altering the activity of CDK2 and the expression of p21 and cyclin E. Birth Defects Res B Dev Reprod Toxicol. 2005;74:233–42.
Zhang JY, et al. Secalonic acid D induced leukemia cell apoptosis and cell cycle arrest of G(1) with involvement of GSK-3beta/beta-catenin/c-Myc pathway. Cell Cycle. 2009;8:2444–50.
Hu YP, et al. Secalonic acid D reduced the percentage of side populations by down-regulating the expression of ABCG2. Biochem Pharmacol. 2013;85:1619–25.
Liao G, et al. The cell toxicity effect of secalonic acid D on GH3 cells and the related mechanisms. Oncol Rep. 2010;23:387–95.
Guru SK, et al. Secalonic acid-D represses HIF1α/VEGF-mediated angiogenesis by regulating the Akt/mTor/p70S6K signaling cascade. Cancer Res. 2015;75:2886–96.
Chen L, et al. Secalonic acids H and I, two new secondary metabolites from the marine-derived fungus penicillium oxalicum. Heterocycles. 2017;94:1766–74.
Ferreira CG, Tolis C, Giaccone G. P53 and chemosensitivity. Ann Oncol. 1999;10:1011–21.
Brady CA, Attardi LD. P53 at a glance. J Cell Sci. 2010;123:2527–32.
Chen L, et al. The marine fungal metabolite, dicitrinone B, induces A375 cell apoptosis through the ROS-related caspase pathway. Mar Drugs. 2014;12:1939–58.
Dong H, et al. Up12, a novel ursolic acid derivative with potential for targeting multiple signaling pathways in hepatocellular carcinoma. Biochem Pharmacol. 2015;93:151–62.
Wang J, et al. Synergism of ursolic acid derivative US597 with 2-deoxy-d-glucose to preferentially induce tumor cell death by dual-targeting of apoptosis and glycolysis. Sci Rep. 2014;4:5006.
Kunnumakkara AB, et al. Curcumin sensitizes human colorectal cancer to capecitabine by modulation of cyclin D1, COX-2, MMP-9, VEGF and CXCR4 expression in an orthotopic mouse model. Int J Cancer. 2009;125:2187–97.
Ma CY, et al. Butein inhibits the migration and invasion of SK-HEP-1 human hepatocarcinoma cells through suppressing the ERK, JNK, p38, and uPA signaling multiple pathways. J Agric Food Chem. 2011;59:9032–8.
Dai C, Gu W. P53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–36.
Tapia N, Scholer HR. P53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med. 2010;207:2045–8.
El-Elimat T, et al. Biosynthetically distinct cytotoxic polyketides from setophoma terrestris. Eur J Org Chem. 2015;109–21.
Qin T, Porco JA. Total syntheses of secalonic acids A and D. Angew Chem Int Ed Engl. 2014;53:3107–10.
Scott DB. Toxigenic fungi isolated from cereal and legume products. Mycopathol Mycol Appl. 1965;25:213–22.
Carlton WW, Tuite J, Mislivec P. Investigations of the toxic effects in mice of certain species of penicillium. Toxicol Appl Pharmacol. 1968;13:372–87.
Hanumegowda UM, Judy BM, Welshons WV, Reddy CS. Selective inhibition of murine palatal mesenchymal cell proliferation in vitro by secalonic acid D. Toxicol Sci. 2002;66:159–65.
Dhulipala VC, Hanumegowda UM, Balasubramanian G, Reddy CS. Relevance of the palatal protein kinase a pathway to the pathogenesis of cleft palate by secalonic acid D in mice. Toxicol Appl Pharmacol. 2004;194:270–9.
Bolon B, St. Omer VE. Behavioral and developmental effects in suckling mice following maternal exposure to the mycotoxin secalonic acid D. Pharmacol Biochem Behav. 1989;34:229–36.
Montella PG, Reddy CS. Neurotoxic effects of secalonic acid D in mice during subchronic postnatal exposure. Pharmacol Biochem Behav. 1991;40:241–7.
Chen LM, et al. The studies on the cytotoxicity in vitro, cellular uptake, cell cycle arrest and apoptosis-inducing properties of ruthenium methylimidazole complex [Ru(Melm)4(p-cpip)](2.). J Inorg Biochem. 2016;156:64–74.
Mongre RK, et al. Epigenetic induction of epithelial to mesenchymal transition by LCN2 mediates metastasis and tumorigenesis, which is abrogated by NF-κB inhibitor BRM270 in a xenograft model of lung adenocarcinoma. Int J Oncol. 2016;48:84–98.
Kiraz Y, Adan A, Kartal Yandim M, Baran Y. Major apoptotic mechanisms and genes involved in apoptosis. Tumour Biol. 2016;37:8471–86.
Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin. 2005;37:719–27.
Blazquez S, et al. Caspase-3 and caspase-6 in ductal breast carcinoma: a descriptive study. Histol Histopathol. 2006;21:1321–9.
Wang L, et al. Targeting notch1 signaling pathway positively affects the sensitivity of osteosarcoma to cisplatin by regulating the expression and/or activity of caspase family. Mol Cancer. 2014;13:139.
Chen CH, et al. Protopine, a novel microtubule-stabilizing agent, causes mitotic arrest and apoptotic cell death in human hormone-refractory prostate cancer cell lines. Cancer Lett. 2012;315:1–11.
Galluzzi L, Larochette N, Zamzami N, Kroemer G. Mitochondria as therapeutic targets for cancer chemotherapy. Oncogene. 2006;25:4812–30.
Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33.
Luijsterburg MS, et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–62.
Girardini JE, Marotta C, Del Sal G. Disarming mutant p53 oncogenic function. Pharmacol Res. 2014;79:75–87.
Brito AF, et al. Hepatocellular carcinoma and chemotherapy: the role of p53. Chemotherapy. 2012;58:381–6.
Lozano G. Mouse models of p53 functions. Cold Spring Harb Perspect Biol. 2010;2:a001115.
Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13.
Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–27.
Qin T, Iwata T, Ransom TT, Beutler JA, Porco JA Jr. Syntheses of dimeric tetrahydroxanthones with varied linkages: Investigation of “shapeshifting” properties. J Am Chem Soc. 2015;137:15225–33.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (21102015 and 31201034), the Key Scientific Project of Ministry of Education in Fujian Province (JZ160419), the Fujian Provincial Science and Technology Innovation Joint Project (2017Y9075), the Medical Innovative Project of Fujian Province (2015-CXB-5), the Natural Science Foundation of Fujian Province (2017J01855, 2017J01179, 2016J01733, and 2014J01301), the Leading Project Foundation of Fujian Province (2018Y0015), “Young Top Creative Talents” of the Second Batch Special Support “Double Hundred Plan” of Fujian Province, and the National Clinical Key Specialty Construction Program of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chen, L., Li, YP., Li, XX. et al. Isolation of 4,4′-bond secalonic acid D from the marine-derived fungus Penicillium oxalicum with inhibitory property against hepatocellular carcinoma. J Antibiot 72, 34–44 (2019). https://doi.org/10.1038/s41429-018-0104-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-018-0104-5